Ribosome Binding Site/Cat page
| RBS part table RBS part icon: |
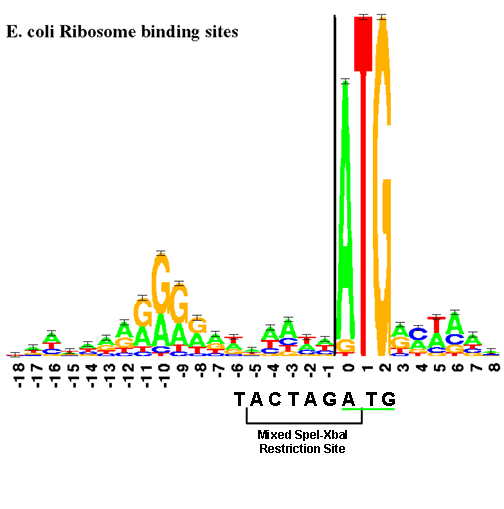
Ribosome Binding Sites
The RBS controls the accuracy and efficiency with which the translation of mRNA begins. A ribosome binding site (RBS) is a segment of the 5' (upstream) part of an mRNA molecule that binds to the ribosome to position the message correctly for translation initiation. In bacteria, translation initiation almost always requires both an RBS sequence and a start codon. In the registry, protein coding sequences begin with the start codon. So, if you want to build a BioBrick system that produces a protein, you need to pick an RBS part and put it upstream of the protein coding sequence you want to translate.
Note that an RBS is often defined as just that part of the mRNA sequence that binds to the ribosome, however, the surrounding sequence can also affect the translation initiation rate. Consequently, a BioBrick™ RBS part contains the classic RBS but sometimes additional surrounding sequence.
| Name | Description | Efficiency * | Length |
|---|---|---|---|
| BBa_B0029 | 1 bp deletion from B0031 | 15 | |
| BBa_B0030 | RBS.1 (strong) -- modified from R. Weiss | 0.6 | 15 |
| BBa_B0031 | RBS.2 (weak) -- derivative of BBa_0030 | 0.07 | 14 |
| BBa_B0032 | RBS.3 (medium) -- derivative of BBa_0030 | 0.3 | 13 |
| BBa_B0033 | RBS.4 (weaker) -- derivative of BBa_0030 | 0.01 | 11 |
| BBa_B0034 | RBS (Elowitz 1999) -- defines RBS efficiency | 1 | 12 |
| BBa_B0035 | RBS (B0030 derivative) | 14 | |
| BBa_B0036 | Specialized RBS | 5 | |
| BBa_B0037 | Specialized RBS | 10 | |
| BBa_B0038 | RBS 1 | 23 | |
| BBa_B0039 | RBS 2 | 22 | |
| BBa_B0041 | RBS 3 | 20 | |
| BBa_B0064 | Tuned RBS for Q04401 | 12 | |
| BBa_B0070 | Specialized RBS modified from that of B0036 (Brink et al.) | 16 | |
| BBa_B0071 | Specialized RBS | 12 | |
| BBa_B0072 | orthogonal RBS | 6 | |
| BBa_B0073 | Specialized RBS | 9 | |
| BBa_B0074 | Orthogonal RBS | 11 | |
| BBa_B0075 | Orthogonal RBS | 8 | |
| BBa_B0076 | Specialized RBS | 7 | |
| BBa_B0077 | Specialized RBS | 10 | |
| BBa_B0078 | Specialized RBS | 12 | |
| BBa_B2001 | T7 RBS 0.3 | 20 | |
| BBa_B2002 | T7 RBS 0.4 | 20 | |
| BBa_B2003 | T7 RBS 0.5 | 20 | |
| BBa_B2017 | 2.8 RBS from T7 | 20 | |
| BBa_B2022 | 4B RBS from T7 | 20 | |
| BBa_B2040 | 8 RBS from T7 | 20 | |
| BBa_I11010 | RBS | 11 | |
| BBa_I20298 | RBS from T7 gene 4 | 20 | |
| BBa_I20299 | RBS from T7 gene 4, optimal aligned spacing | 16 | |
| BBa_I723012 | Estimated RBS for DntR | 24 | |
| BBa_I723014 | Estimated RBS for DntA | 25 | |
| BBa_I723019 | RBS for XylR | 20 | |
| BBa_I742130 | Reverse RBS | 10 | |
| BBa_I742145 | sam5 native rbs | 16 | |
| BBa_I742146 | sam8 native rbs | 14 | |
| BBa_I742150 | crtE native rbs | 42 | |
| BBa_I742153 | crtY native rbs | 19 | |
| BBa_I742156 | crtZ native rbs | 16 | |
| BBa_I742159 | crtI native rbs | 22 | |
| BBa_I742163 | native ftsK rbs | 26 | |
| BBa_J100112 | BD24 C.dog cloned into J100110 | 191 | |
| BBa_J100296 | rClone Red Version 2 with RBS 2.0: Device for GGA Cloning and Testing RBS elements and Riboswitches | ||
| BBa_J100297 | rClone Red Version 1.5 with RBS 2.0: Device for GGA Cloning and Testing RBS elements and Riboswitche | ||
| BBa_J100331 | Mutated RBS | 11 | |
| BBa_J100335 | Attempt at the Creation of a Strong RBS | 41 | |
| BBa_J100339 | Mutated RBS | 11 | |
| BBa_J100426 | BBa_J119028 w/o first 33bp | 55 | |
| BBa_J100439 | Lots of As | 19 | |
| BBa_J100440 | WC | 11 | |
| BBa_J100449 | Testing non-Watson and Crick base pairing within the RBS sequence | 19 | |
| BBa_J100450 | This is to test a promoter | ||
| BBa_J100453 | Examining the effect of a single base mutation in a RBS sequence on RFP production | 36 | |
| BBa_J100454 | RBS non Watson-Crick pairing (pairing A-C) | 19 | |
| BBa_J100457 | RBS for 16s rClone | 20 | |
| BBa_J100458 | Even More As | 19 | |
| BBa_J100461 | Our RBS is built off of principles of the 2D shape of mRNA. | 31 | |
| BBa_J100462 | The Hy5 | 36 | |
| BBa_J100466 | E.coli Plasmid Ribosomal Binding Sequence | 35 | |
| BBa_J100467 | A new RBS in E. coli to code for mScarlet protein | 23 | |
| BBa_J100470 | Examining the effect of a single base mutation in the sequence prior to RBS on RFP production | 36 | |
| BBa_J100471 | RBS non-Watson Crick base pairing (A-A) | 11 | |
| BBa_J107130 | RC1 RBS (RBS) | 29 | |
| BBa_J107131 | RC2 RBS (RBS) | 29 | |
| BBa_J107132 | RC3 RBS (RBS) | 29 | |
| BBa_J119053 | J23100 Promoter with BD18 RBS w/ leader and RFP reporter | 870 | |
| BBa_J119390 | Collection of RBSs from N6 rClone Red Library | 8 | |
| BBa_J119391 | Collection of RBSs from N8 rClone Red Library | 8 | |
| BBa_J119392 | Collection of Bicistronic RBSs from N6 rClone Red Library | 8 | |
| BBa_J119393 | Collection of Bicistronic RBSs from N8 rClone Red Library | 8 | |
| BBa_J119419 | BCD1 | 802 | |
| BBa_J119420 | BCD9 | 802 | |
| BBa_J119421 | BCD10 | 802 | |
| BBa_J119422 | BCD17 | 802 | |
| BBa_J119423 | BCD18 | 802 | |
| BBa_J119424 | BCD20 | 802 | |
| BBa_J119425 | BCD24 | 802 | |
| BBa_J119428 | BCD16 | 802 | |
| BBa_J119429 | BCD22 | 802 | |
| BBa_J119430 | BCD6 | 802 | |
| BBa_J119431 | BCD14 | 802 | |
| BBa_J119432 | BCD6 with TA | 804 | |
| BBa_J119433 | BCD10 with TA | 804 | |
| BBa_J119434 | BCD14 with TA | 804 | |
| BBa_J119435 | BCD16 with TA | 804 | |
| BBa_J119436 | BCD20 with TA | 804 | |
| BBa_J119437 | BCD22 with TA | 804 | |
| BBa_J146000 | Consensus RBS | 37 | |
| BBa_J15001 | strong synthetic E. coli ribosome binding site with SacI site. | 10 | |
| BBa_J176012 | Kozak | 18 | |
| BBa_J18912 | T7 promoter + RBS + START | 83 | |
| BBa_J23080 | b0034 derived RBS to match spacing of J01122 | 10 | |
| BBa_J26002 | Mario Binding Site | 6 | |
| BBa_J29048 | RBS | 13 | |
| BBa_J329998 | Short Version of RBS with Predicted TIR of 99 au | 14 | |
| BBa_J329999 | RBS with Predicted TIR of 99 au for BBa_C0061 | 31 | |
| BBa_J331018 | RBS for hsp90 | 56 | |
| BBa_J34801 | ribosome binding site | 12 | |
| BBa_J34803 | ribosome binding site | 13 | |
| BBa_J34810 | ribosome binding site | 8 | |
| BBa_J364100 | Bicistronic Design Element, Number 2 (BCD2) | 84 | |
| BBa_J428032 | BBa_B0030_m0 | 24 | |
| BBa_J428034 | BCD1 | 84 | |
| BBa_J428037 | BBa_B0031_m0 | 23 | |
| BBa_J428038 | BBa_B0034_m1 | 22 | |
| BBa_J428039 | SpoVG | 21 | |
| BBa_J433003 | 5UTR13 5prime UTR | 22 | |
| BBa_J433004 | uORFs (1x weak) | 22 | |
| BBa_J433005 | uORFs (1x) | 22 | |
| BBa_J433006 | uORFs (4x) | 58 | |
| BBa_J434008 | BCD13 | 84 | |
| BBa_J434141 | pET RBS for OYC dropout | 21 | |
| BBa_J435302 | BN1_Riboj54_F1 | 114 | |
| BBa_J435303 | BN1_BCD2_pelB | 152 | |
| BBa_J435305 | BC_BCD12 | 84 | |
| BBa_J435322 | BN1_BCD12 | 89 | |
| BBa_J435332 | BN1_BCD23 | 89 | |
| BBa_J435342 | BN1_BCD2_His | 119 | |
| BBa_J435352 | BN1_RiboJ_B34_His | 133 | |
| BBa_J435354 | BN2_RiboJ_B34 | 101 | |
| BBa_J435362 | BN1_BCD2_DsbA | 725 | |
| BBa_J435364 | BC_B0034 | 21 | |
| BBa_J435372 | BN1_BCD2_OmpT | 167 | |
| BBa_J435374 | BC_B0032 | 22 | |
| BBa_J435385 | BC_BCD23 | 84 | |
| BBa_J435386 | BC_RiboJ_B34 | 98 | |
| BBa_J435387 | BC_RiboJ_RBS64 | 100 | |
| BBa_J44001 | Reverse RBS (RBSrev) -- corresponds to BBa_B0030 | n/a | 15 |
| BBa_J56013 | Rbs2 ribosome binding site | 19 | |
| BBa_J56016 | Rbs-orig - ribosome binding site | 21 | |
| BBa_J59001 | tuba | 34 | |
| BBa_J61100 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61101 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61102 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61103 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61104 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61105 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61106 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61107 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61108 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61109 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61110 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61111 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61112 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61113 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61114 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61115 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61116 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61117 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61118 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61119 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61120 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61121 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61122 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61123 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61124 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61125 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61126 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61127 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61128 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61129 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61130 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61131 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61132 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61133 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61134 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61135 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61136 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61137 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61138 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61139 | Ribosome Binding Site Family Member | 12 | |
| BBa_J61140 | {rbs1} Library Member in BBb | 15 | |
| BBa_J61141 | {rbs1} Library Member in BBb | 15 | |
| BBa_J61142 | {rbs1} Library Member in BBb | 15 | |
| BBa_J61143 | {rbs1} Library Member in BBb | 15 | |
| BBa_J61144 | {rbs1} Library Member in BBb | 15 | |
| BBa_J61145 | {rbs1} Library Member in BBb | 15 | |
| BBa_J61146 | {rbs1} Library Member in BBb | 15 | |
| BBa_J61147 | {rbs1} Library Member in BBb | 15 | |
| BBa_J63003 | designed yeast Kozak sequence | n/a | 18 |
| BBa_J64011 | invB RBS | 19 | |
| BBa_J64013 | sipA RBS | 18 | |
| BBa_J64015 | sopA RBS | 16 | |
| BBa_J64018 | invJ RBS | 17 | |
| BBa_J64020 | invI RBS | 20 | |
| BBa_J64022 | sicA RBS | 20 | |
| BBa_J64024 | sipC RBS | 15 | |
| BBa_J64026 | sigE RBS | 18 | |
| BBa_J64028 | sopB RBS | 20 | |
| BBa_J64030 | sopD RBS | 17 | |
| BBa_J64031 | sptP RBS | 20 | |
| BBa_J64609 | creD RBS | 22 | |
| BBa_J64807 | RocD RBS | 16 | |
| BBa_J64808 | RocE RBS | 14 | |
| BBa_J64809 | RocF RBS | 14 | |
| BBa_J64907 | creA RBS | 21 | |
| BBa_J64908 | RBS for creC in e coli operon | 24 | |
| BBa_J64968 | RBS on creB | 21 | |
| BBa_J70591 | RFC12 Elowitz RBS | 12 | |
| BBa_J73036 | Ethylene Forming Enzyme coding sequence | 1065 | |
| BBa_J73105 | Ribosome binding site | ||
| BBa_J90000 | Weak RBS | 11 | |
| BBa_J95015 | Strong RBS for Rhodobacter sphaeroides | 12 | |
| BBa_J95016 | Weak RBS for Rhodobacter sphaeroides | 12 | |
| BBa_J95017 | Weaker RBS-GGAGG8 for Rhodobacter sphaeroides | 12 | |
| BBa_J95018 | Medium RBS-GGAGG10 for Rhodobacter sphaeroides | 12 | |
| BBa_J95019 | Medium RBS-GGAGG11 for Rhodobacter sphaeroides | 12 | |
| BBa_J95021 | RBS for Rhodobacter sphaeroides | 12 | |
| BBa_J95028 | RBS for Rhodobacter sphaeroides | 12 | |
| BBa_J96040 | IRES | 504 | |
| BBa_J96043 | PS3 | 50 | |
| BBa_J96044 | PS4 | 48 | |
| BBa_K082000 | reconstruct strong RBS | 17 | |
| BBa_K082001 | RBS medium | 15 | |
| BBa_K082002 | RBS defines RBS efficiency | 14 | |
| BBa_K090505 | ''Bacillus subtilis'' consensus RBS | 11 | |
| BBa_K090506 | ''Bacillus subtilis'' weak RBS | 11 | |
| BBa_K1017202 | Regulation RBS-2 | 21 | |
| BBa_K103015 | consensus RBS | 6 | |
| BBa_K104003 | RBS_spaR | 9 | |
| BBa_K104005 | RBS_spaK | 9 | |
| BBa_K104009 | RBS_gfp | 8 | |
| BBa_K1045010 | RBS BBa_B0034 with inversed Pre- and Suffix | 12 | |
| BBa_K1074015 | RBS for Pgrac | 16 | |
| BBa_K1074016 | RBS for Pctc | 18 | |
| BBa_K1084101 | SD2 | 33 | |
| BBa_K1084102 | SD4 | 33 | |
| BBa_K1084103 | SD6 | 33 | |
| BBa_K1084104 | SD8 | 33 | |
| BBa_K1088055 | RBS from pKT25 | 19 | |
| BBa_K1100000 | ALeader | 75 | |
| BBa_K1100001 | ALeaderT:The First Artificial Triphase Riboswitch | 87 | |
| BBa_K1100014 | ALeaderT (with insulator) | 106 | |
| BBa_K1100071 | RNA-IN S1 | 32 | |
| BBa_K1100072 | RNA-IN S4 | 32 | |
| BBa_K1100073 | RNA-IN S5 | 32 | |
| BBa_K1100074 | RNA-IN S31 | 32 | |
| BBa_K1100075 | RNA-IN S34 | 34 | |
| BBa_K1100076 | RNA-IN S49 | 34 | |
| BBa_K1114100 | MoClo format of a modified version of BBa_B0030 with BC fusion sites. | 32 | |
| BBa_K1114101 | MoClo format of a modified version of BBa_B0031 with BC fusion sites. | 31 | |
| BBa_K1114102 | MoClo format of a modified version of BBa_B0032 with BC fusion sites | 30 | |
| BBa_K1114103 | MoClo formatted version of B0033 with BC fusion sites | 28 | |
| BBa_K1114104 | MoClo formatted version of B0034 with BC fusion sites | 29 | |
| BBa_K1114106 | Bicistronic Design RBS 1, MoClo Format with BC fusion sites | 92 | |
| BBa_K1114107 | Bicistronic Design RBS 2, MoClo Format with BC fusion sites | 92 | |
| BBa_K1114108 | Bicistronic design RBS 8, MoClo Format with BC fusion sites | 92 | |
| BBa_K1114109 | Bicistronic design RBS 12, MoClo Format with BC fusion sites | 92 | |
| BBa_K1114110 | Bicistronic design RBS 13, MoClo Format with BC fusion sites | 92 | |
| BBa_K1114111 | Bicistronic design RBS 16, MoClo Format with BC fusion sites | 92 | |
| BBa_K1123010 | Ribosoom Binding Site | 22 | |
| BBa_K1130003 | L. plantarum RBS | 33 | |
| BBa_K1145003 | A ribosome switch which can be opened by atrazine. | 87 | |
| BBa_K1159300 | Polioviral Internal Ribosome Entry Site (IRES) for polycistronic expression in eukaryotes | 627 | |
| BBa_K118012 | Synthetic ribosome binding site added by Son of Babel procedure | 4 | |
| BBa_K121016 | Tokyo Standard prefix suffix RBS | 64 | |
| BBa_K1231002 | Lpp-RBS is a constitutive promoter | 90 | |
| BBa_K1323016 | Translational initiation region (RBS) for Staphylococcus aureus | 28 | |
| BBa_K1323022 | Ribosome binding site from the sodA Staphylococcus aureus gene | 18 | |
| BBa_K1341023 | LTT-rbs | 13 | |
| BBa_K1351028 | LMU Bacillus RBS collection 1 | 11 | |
| BBa_K1351029 | LMU Bacillus RBS collection 2 | 11 | |
| BBa_K1351030 | LMU Bacillus RBS collection 3 | 11 | |
| BBa_K1351031 | LMU Bacillus RBS collection 4 | 11 | |
| BBa_K1351032 | LMU Bacillus RBS collection 5 | 11 | |
| BBa_K1351033 | LMU Bacillus RBS collection 6 | 11 | |
| BBa_K1351034 | LMU Bacillus RBS collection 7 | 11 | |
| BBa_K1362090 | strong T7 RBS | 29 | |
| BBa_K1379015 | B0032 derivate for RBS used in Lock 3C | 12 | |
| BBa_K1403008 | sRBS | 35 | |
| BBa_K1403010 | RBS for agaA in E coli | 35 | |
| BBa_K1403011 | sRBS | 35 | |
| BBa_K1403020 | sRBS | 35 | |
| BBa_K1419003 | Universal riboregulatable RBS | 39 | |
| BBa_K143020 | GsiB ribosome binding site (RBS) for B. subtilis | 11 | |
| BBa_K143021 | SpoVG ribosome binding site (RBS) for B. subtilis | 12 | |
| BBa_K1442000 | Test | 28 | |
| BBa_K1442008 | EMCV IRES | ||
| BBa_K1442023 | EMCV IRES | 625 | |
| BBa_K1442024 | NKRF IRES | 667 | |
| BBa_K1442109 | EMCV IRES with forward GFP | 1432 | |
| BBa_K1442110 | NKRF IRES with forward GFP | 1498 | |
| BBa_K1453790 | try | ||
| BBa_K1453791 | try | ||
| BBa_K1461000 | crRBS | 52 | |
| BBa_K1463100 | GvpA, K737016, RBS | 8 | |
| BBa_K1463560 | Reverse RBS | 10 | |
| BBa_K1471011 | RBS (Arabidopsis Thaliana). | 11 | |
| BBa_K1479003 | Just for upload test | ||
| BBa_K1479004 | Just for upload test | 431 | |
| BBa_K1479006 | Just for upload test | 32 | |
| BBa_K1479007 | Just for upload test | 32 | |
| BBa_K1479015 | Just for test | 16 | |
| BBa_K150005 | ribosome binding site of pTrc99a | 13 | |
| BBa_K1529948 | abe | 24 | |
| BBa_K1529997 | aa | 12 | |
| BBa_K1583008 | RBS | 12 | |
| BBa_K1583058 | RBS | 12 | |
| BBa_K1583061 | RBS | 12 | |
| BBa_K1605002 | nifZ RBS | 15 | |
| BBa_K1605004 | nifE RBS | 15 | |
| BBa_K1605006 | nifH RBS | 15 | |
| BBa_K1605008 | Ptet mRFP nifS RBS | 15 | |
| BBa_K1605010 | nifK RBS | 15 | |
| BBa_K1605012 | nifB RBS | 15 | |
| BBa_K1605014 | nifD RBS | 15 | |
| BBa_K1605016 | nifN RBS | 15 | |
| BBa_K1605018 | hesB RBS | 16 | |
| BBa_K1605020 | hesA RBS | 15 | |
| BBa_K1605022 | cysE2 RBS | 15 | |
| BBa_K1605024 | nifV RBS | 15 | |
| BBa_K1616011 | RBS reversed | 14 | |
| BBa_K1624003 | RBS | 7 | |
| BBa_K1638001 | RBS from pKT25 and pUT18 | 19 | |
| BBa_K1638017 | RBS from pKT25 and pUT18C with consensus Shine-Dalgarno | 19 | |
| BBa_K1642004 | SD sequence of cpcB | 20 | |
| BBa_K165002 | Kozak sequence (yeast RBS) | 18 | |
| BBa_K1650034 | RBS | 12 | |
| BBa_K1676001 | RBS Mutant 1AC | 12 | |
| BBa_K1676002 | RBS Mutant 1AG | 12 | |
| BBa_K1676003 | RBS Mutant 1AT | 12 | |
| BBa_K1676004 | RBS Mutant 2AC | 12 | |
| BBa_K1676005 | RBS Mutant 2AG | 12 | |
| BBa_K1676006 | RBS Mutant 2AT | 12 | |
| BBa_K1676007 | RBS Mutant 3AC | 12 | |
| BBa_K1676008 | RBS Mutant 3AG | 12 | |
| BBa_K1676009 | RBS Mutant 3AT | 12 | |
| BBa_K1676010 | RBS Mutant 4GA | 12 | |
| BBa_K1676011 | RBS Mutant 4GC | 12 | |
| BBa_K1676012 | RBS Mutant 4GT | 12 | |
| BBa_K1676013 | RBS Mutant 5AC | 12 | |
| BBa_K1676014 | RBS Mutant 5AG | 12 | |
| BBa_K1676015 | RBS Mutant 5AT | 12 | |
| BBa_K1676016 | RBS Mutant 6GA | 12 | |
| BBa_K1676017 | RBS Mutant 6GC | 12 | |
| BBa_K1676018 | RBS Mutant 6GT | 12 | |
| BBa_K1676019 | RBS Mutant 7GA | 12 | |
| BBa_K1676020 | RBS Mutant 7GC | 12 | |
| BBa_K1676021 | RBS Mutant 7GT | 12 | |
| BBa_K1676022 | RBS Mutant 8AC | 12 | |
| BBa_K1676023 | RBS Mutant 8AG | 12 | |
| BBa_K1676024 | RBS Mutant 8AT | 12 | |
| BBa_K1676025 | RBS Mutant 9GA | 12 | |
| BBa_K1676026 | RBS Mutant 9GC | 12 | |
| BBa_K1676027 | RBS Mutant 9GT | 12 | |
| BBa_K1676028 | RBS Mutant 10AC | 12 | |
| BBa_K1676029 | RBS Mutant 10AG | 12 | |
| BBa_K1676030 | RBS Mutant 10AT | 12 | |
| BBa_K1676031 | RBS Mutant 11AC | 12 | |
| BBa_K1676032 | RBS Mutant 11AG | 12 | |
| BBa_K1676033 | RBS Mutant 11AT | 12 | |
| BBa_K1676034 | RBS Mutant 12AC | 12 | |
| BBa_K1676035 | RBS Mutant 12AG | 12 | |
| BBa_K1676036 | RBS Mutant 12AT | 12 | |
| BBa_K1676073 | Mutant RBS Opt | 12 | |
| BBa_K1705011 | rCHU_1284 | 32 | |
| BBa_K1725301 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725302 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725303 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725304 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725305 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725306 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725307 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725308 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725309 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725310 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725311 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725312 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725313 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725314 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725315 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725316 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725317 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725318 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725319 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725320 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725321 | Member of the RBS library - derived from BBa_0032 | 13 | |
| BBa_K1725322 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725323 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725324 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725325 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725326 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725327 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725328 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725329 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725330 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725331 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1725332 | Member of the RBS library - derived from BBa_B0032 | 13 | |
| BBa_K1759900 | try | ||
| BBa_K1789002 | RBS30 | 15 | |
| BBa_K1799011 | LsrA RBS | 10 | |
| BBa_K1799012 | LsrC RBS | 10 | |
| BBa_K1799013 | LsrD RBS | 10 | |
| BBa_K1799014 | LsrB RBS | 10 | |
| BBa_K1799028 | RBS.2 | 50 | |
| BBa_K1799029 | RBS.4 | 48 | |
| BBa_K1841003 | ribosome binding site for lactobacillus casei | 16 | |
| BBa_K1841012 | rbs (lactobacillus casei) | 12 | |
| BBa_K1875001 | This part contains a Kozak sequence | 9 | |
| BBa_K1893033 | Strong ribosome binding site (RBS) | 20 | |
| BBa_K1895001 | rpoH | 16 | |
| BBa_K1897031 | B0033 with flanking Scar sites | 25 | |
| BBa_K1897032 | B0032 with flanking Scar sites | 27 | |
| BBa_K1897034 | B0032-Scar site | 19 | |
| BBa_K1897036 | B0033-Scar site | 17 | |
| BBa_K1906010 | Ribothermometer A1 | 42 | |
| BBa_K1906011 | Ribothermometer JB2-B1 | 42 | |
| BBa_K1906012 | RNAT Mut | 42 | |
| BBa_K1906013 | Ribothermometer JB2-C6 | 42 | |
| BBa_K1906014 | Ribothermometer JB1-A8 | 42 | |
| BBa_K1906015 | Ribothermometer JB1-B2 | 42 | |
| BBa_K1906016 | Ribothermometer JB1_B5 | 37 | |
| BBa_K1906017 | Ribothermometer JB2-B6 | 42 | |
| BBa_K1906018 | Ribothermometer JB1-G3 | 42 | |
| BBa_K1906019 | Ribothermometer JB1-G4 | 42 | |
| BBa_K1906020 | ribothermometer modified A1 to work as positive cpntrol | 42 | |
| BBa_K1906021 | ribothermometer | ||
| BBa_K1925000 | sdasd | ||
| BBa_K1928006 | jdklajkfl | ||
| BBa_K1968002 | Synechocystis RBS* Phytobrick | 23 | |
| BBa_K1992000 | Tar native RBS | 24 | |
| BBa_K1993016 | IRES | 588 | |
| BBa_K1994002 | RBS 173k TIRv1 | 30 | |
| BBa_K1994004 | RBS 28.4k_NEQ RBSCv1.1 | 47 | |
| BBa_K1994007 | synRBS 9675 TIRv(1 or 2) | 30 | |
| BBa_K2018026 | Ribosomal binding site for hlyB | 71 | |
| BBa_K2018028 | RBS for hlyD | 18 | |
| BBa_K2018032 | RBS for panK | 21 | |
| BBa_K2027073 | (pMS-AFa-DEa) pAKP444 RBS 1 extraction | 28 | |
| BBa_K2027074 | (pMS-AFa-DEa) pAKP444 RBS 2 extraction | 26 | |
| BBa_K2042003 | RBS | 7 | |
| BBa_K2066510 | Strong RBS from Lou et. al 2012 | 12 | |
| BBa_K2066515 | RBS2 (weak) | 12 | |
| BBa_K2066526 | B0034 with scar TACTAG | ||
| BBa_K2066527 | B0034 with scar TACTAG | 18 | |
| BBa_K2066549 | Synthetic Enhancer Project: RBS | 1 | |
| BBa_K208100 | Rhodobacter sphaeroides RBS | 21 | |
| BBa_K208101 | Rhodobacter sphaeroides RBS | 21 | |
| BBa_K208102 | Rhodobacter sphaeroides RBS | 21 | |
| BBa_K208103 | Rhodobacter sphaeroides RBS | 21 | |
| BBa_K208104 | Rhodobacter sphaeroides RBS | 21 | |
| BBa_K208105 | Rhodobacter sphaeroides RBS | 21 | |
| BBa_K208106 | Rhodobacter sphaeroides RBS | 21 | |
| BBa_K208107 | Rhodobacter sphaeroides RBS | 21 | |
| BBa_K208108 | Rhodobacter sphaeroides RBS | 21 | |
| BBa_K2116019 | RBS designed for sfgfp | 35 | |
| BBa_K2116020 | RBS for mNectarine | 32 | |
| BBa_K2116031 | RBS for sfGFP | 41 | |
| BBa_K2116033 | RBS_esaR | 17 | |
| BBa_K2116094 | Salis RBS for BB assembly | 32 | |
| BBa_K2116999 | RBS for EsaR | 17 | |
| BBa_K2138000 | RNA thermometer and RBS (40C) | 2187 | |
| BBa_K2138001 | RNA thermometer and RBS (39C) | 2283 | |
| BBa_K2138002 | RNA thermometer and RBS (33C) | 2175 | |
| BBa_K2182004 | RBS for BBa_K2182003 | 22 | |
| BBa_K2200999 | 1H2IU3G | ||
| BBa_K2205098 | B0034 (Reversed) | 12 | |
| BBa_K2232030 | RBS of B.subtilis | 26 | |
| BBa_K2241000 | 233? | ||
| BBa_K2284026 | strong_RBS_including_spacer_Notts | 18 | |
| BBa_K2285021 | RBS on sfGFP | 24 | |
| BBa_K2290004 | Ribosome Binding Site | 14 | |
| BBa_K2302012 | Ribosome Binding Sites | 18 | |
| BBa_K2306014 | Efficient RBS from bacteriophage T7 gene 10 and Shine Dalgarno | 30 | |
| BBa_K2328014 | RBS I for E.coli | 24 | |
| BBa_K2328015 | RBS II | 25 | |
| BBa_K2328016 | RBS III for B.longum (with ATG) | 26 | |
| BBa_K2333008 | mScarlet-I no stop codon | 696 | |
| BBa_K2340008 | Guide RNA construct of human Pkp4 (2) | 535 | |
| BBa_K234001 | -- No description -- | ||
| BBa_K234090 | -- No description -- | ||
| BBa_K2355219 | Strong promoter with RFC[25] scars and a startcodon | 30 | |
| BBa_K2378010 | aaaaaaaaaaaaaaaaaa | ||
| BBa_K2380024 | RBS B0034 with BpiI recognition site | 18 | |
| BBa_K2387051 | RBS with Scars | 28 | |
| BBa_K2389004 | RBS for pT8O | 15 | |
| BBa_K2390002 | S. Mutans B-9 Responsive Riboswitch | 223 | |
| BBa_K239008 | RBS from RpoS | 567 | |
| BBa_K2406216 | MoClo RBS B0034 | 21 | |
| BBa_K2417009 | Extended ribosome binding site | 33 | |
| BBa_K2418005 | Extended Ribosome Binding Site (RBS) | 45 | |
| BBa_K2442210 | Wild Type RBS from P.fluorescens | 83 | |
| BBa_K2448000 | B0034 with scars AGGA and AGATCA | 22 | |
| BBa_K2448002 | B0034 like RBS with scars AATT and CAGCT | 20 | |
| BBa_K2448019 | RBS (pET like) | 25 | |
| BBa_K2448020 | B0034 with scars TAC and CAT | 18 | |
| BBa_K2448039 | B0034 with scars GTAC and TACCAT | 22 | |
| BBa_K2448049 | B0034 with scars GTAC and CTCGAGG | 23 | |
| BBa_K2448060 | B0034 with scars AAATGGAGGA and AGATCA | 28 | |
| BBa_K2449033 | SDparM | 7 | |
| BBa_K2462001 | RBS | 10 | |
| BBa_K2462002 | RBS | 10 | |
| BBa_K2491021 | Promoter with RBS (100) | 47 | |
| BBa_K2491022 | Promoter with RBS (101) | 47 | |
| BBa_K2491023 | Promoter with RBS (110) | 47 | |
| BBa_K252015 | f | ||
| BBa_K2541001 | Heat-inducible RNA-based thermosensor-1 | 23 | |
| BBa_K2541002 | Heat-inducible RNA-based thermosensor-2 | 33 | |
| BBa_K2541003 | Heat-inducible RNA-based thermosensor-3 | 29 | |
| BBa_K2541004 | Heat-inducible RNA-based thermosensor-4 | 35 | |
| BBa_K2541005 | Heat-inducible RNA-based thermosensor-5 | 27 | |
| BBa_K2541006 | Heat-inducible RNA-based thermosensor-6 | 27 | |
| BBa_K2541007 | Heat-inducible RNA-based thermosensor-7 | 37 | |
| BBa_K2541008 | Heat-inducible RNA-based thermosensor-8 | 35 | |
| BBa_K2541009 | Heat-inducible RNA-based thermosensor-9 | 41 | |
| BBa_K2541010 | Heat-inducible RNA-based thermosensor-10 | 30 | |
| BBa_K2541011 | Heat-inducible RNA-based thermosensor-11 | 39 | |
| BBa_K2541012 | Heat-inducible RNA-based thermosensor-12 | 39 | |
| BBa_K2541013 | Heat-inducible RNA-based thermosensor-13 | 38 | |
| BBa_K2541014 | Heat-inducible RNA-based thermosensor-14 | 27 | |
| BBa_K2541015 | Heat-inducible RNA-based thermosensor-15 | 27 | |
| BBa_K2541016 | Heat-inducible RNA-based thermosensor-16 | 24 | |
| BBa_K2541017 | Heat-inducible RNA-based thermosensor-17 | 24 | |
| BBa_K2541018 | Heat-inducible RNA-based thermosensor-18 | 27 | |
| BBa_K2541019 | Heat-inducible RNA-based thermosensor-19 | 28 | |
| BBa_K2541020 | Heat-inducible RNA-based thermosensor-20 | 35 | |
| BBa_K2541021 | Heat-inducible RNA-based thermosensor-21 | 35 | |
| BBa_K2541025 | Heat-inducible RNA-based thermosensor-25 | 41 | |
| BBa_K2541026 | Heat-inducible RNA-based thermosensor-26 | 30 | |
| BBa_K2541027 | Heat-inducible RNA-based thermosensor-27 | 29 | |
| BBa_K2541028 | Heat-inducible RNA-based thermosensor-28 | 39 | |
| BBa_K2541029 | Heat-inducible RNA-based thermosensor-29 | 34 | |
| BBa_K2541030 | Heat-inducible RNA-based thermosensor-30 | 33 | |
| BBa_K2541031 | Heat-inducible RNA-based thermosensor-31 | 39 | |
| BBa_K2541032 | Heat-inducible RNA-based thermosensor-32 | 39 | |
| BBa_K2541033 | Heat-inducible RNA-based thermosensor-33 | 38 | |
| BBa_K2541034 | Heat-inducible RNA-based thermosensor-34 | 39 | |
| BBa_K2541035 | Heat-inducible RNA-based thermosensor-35 | 39 | |
| BBa_K2541036 | Heat-inducible RNA-based thermosensor-36 | 27 | |
| BBa_K2541037 | Heat-inducible RNA-based thermosensor-37 | 24 | |
| BBa_K2541038 | Heat-inducible RNA-based thermosensor-38 | 24 | |
| BBa_K2541039 | Heat-inducible RNA-based thermosensor-39 | 28 | |
| BBa_K2541040 | Heat-inducible RNA-based thermosensor-40 | 28 | |
| BBa_K2541041 | Heat-inducible RNA-based thermosensor-41 | 28 | |
| BBa_K2541042 | Heat-inducible RNA-based thermosensor-42 | 24 | |
| BBa_K2541043 | Heat-inducible RNA-based thermosensor-43 | 34 | |
| BBa_K2541044 | Heat-inducible RNA-based thermosensor-44 | 29 | |
| BBa_K2541045 | Heat-inducible RNA-based thermosensor-45 | 31 | |
| BBa_K2541046 | Heat-inducible RNA-based thermosensor-46 | 35 | |
| BBa_K2541047 | Heat-inducible RNA-based thermosensor-47 | 41 | |
| BBa_K2541048 | Heat-inducible RNA-based thermosensor-48 | 30 | |
| BBa_K2541049 | Heat-inducible RNA-based thermosensor-49 | 34 | |
| BBa_K2541050 | Heat-inducible RNA-based thermosensor-50 | 40 | |
| BBa_K2541051 | Heat-inducible RNA-based thermosensor-51 | 39 | |
| BBa_K2541101 | Heat-repressible RNA-based thermosensor-1 | 69 | |
| BBa_K2541102 | Heat-repressible RNA-based thermosensor-2 | 70 | |
| BBa_K2541103 | Heat-repressible RNA-based thermosensor-3 | 74 | |
| BBa_K2541104 | Heat-repressible RNA-based thermosensor-4 | 75 | |
| BBa_K2541105 | Heat-repressible RNA-based thermosensor-5 | 79 | |
| BBa_K2541106 | Heat-repressible RNA-based thermosensor-6 | 80 | |
| BBa_K2541108 | Heat-repressible RNA-based thermosensor-8 | 74 | |
| BBa_K2541109 | Heat-repressible RNA-based thermosensor-9 | 78 | |
| BBa_K2541110 | Heat-repressible RNA-based thermosensor-10 | 60 | |
| BBa_K2541111 | Heat-repressible RNA-based thermosensor-11 | 60 | |
| BBa_K2541112 | Heat-repressible RNA-based thermosensor-12 | 61 | |
| BBa_K2541113 | Heat-repressible RNA-based thermosensor-13 | 65 | |
| BBa_K2541114 | Heat-repressible RNA-based thermosensor-14 | 66 | |
| BBa_K2541115 | Heat-repressible RNA-based thermosensor-15 | 70 | |
| BBa_K2541116 | Heat-repressible RNA-based thermosensor-16 | 61 | |
| BBa_K2541117 | Heat-repressible RNA-based thermosensor-17 | 65 | |
| BBa_K2541118 | Heat-repressible RNA-based thermosensor-18 | 54 | |
| BBa_K2541119 | Heat-repressible RNA-based thermosensor-19 | 53 | |
| BBa_K2541120 | Heat-repressible RNA-based thermosensor-20 | 54 | |
| BBa_K2541121 | Heat-repressible RNA-based thermosensor-21 | 58 | |
| BBa_K2541122 | Heat-repressible RNA-based thermosensor-22 | 53 | |
| BBa_K2541123 | Heat-repressible RNA-based thermosensor-23 | 59 | |
| BBa_K2541201 | Cold-repressible RNA-based thermosensor-1 | 69 | |
| BBa_K2541202 | Cold-repressible RNA-based thermosensor-2 | 69 | |
| BBa_K2541203 | Cold-repressible RNA-based thermosensor-3 | 69 | |
| BBa_K2541204 | Cold-repressible RNA-based thermosensor-4 | 69 | |
| BBa_K2541205 | Cold-repressible RNA-based thermosensor-5 | 69 | |
| BBa_K2541206 | Cold-repressible RNA-based thermosensor-6 | 69 | |
| BBa_K2541207 | Cold-repressible RNA-based thermosensor-7 | 69 | |
| BBa_K2541208 | Cold-repressible RNA-based thermosensor-8 | 69 | |
| BBa_K2541209 | Cold-repressible RNA-based thermosensor-9 | 69 | |
| BBa_K2541210 | Cold-repressible RNA-based thermosensor-10 | 69 | |
| BBa_K2541301 | Cold-inducible RNA-based thermosensor-1 | 169 | |
| BBa_K2541302 | Cold-inducible RNA-based thermosensor-2 | 169 | |
| BBa_K2541303 | Cold-inducible RNA-based thermosensor-3 | 169 | |
| BBa_K2541304 | Cold-inducible RNA-based thermosensor-4 | 169 | |
| BBa_K2541305 | Cold-inducible RNA-based thermosensor-5 | 169 | |
| BBa_K2541306 | Cold-inducible RNA-based thermosensor-6 | 163 | |
| BBa_K2541307 | Cold-inducible RNA-based thermosensor-7 | 163 | |
| BBa_K2541308 | Cold-inducible RNA-based thermosensor-8 | 163 | |
| BBa_K2550200 | RBS for Toehold Switches | 8 | |
| BBa_K2556021 | RBS from bacteriophage T7 gene 10 | 39 | |
| BBa_K2556022 | RBS | 25 | |
| BBa_K2560008 | Phytobrick version of BBa_B0034 | 21 | |
| BBa_K2560010 | Phytobrick version of BBa_B0032 | 22 | |
| BBa_K2560013 | Phytobrick version of BBa_B0031 | 23 | |
| BBa_K2560016 | Phytobrick version of BBa_B0030 | 24 | |
| BBa_K2560084 | Phytobrick version of RBS Dummy | 24 | |
| BBa_K2586008 | RBS: Ribosome Binding Site | 13 | |
| BBa_K2586010 | Palf4-RBS: Palf4 linked to a RBS | 51 | |
| BBa_K2593005 | itís a strong Ribosome Binding Site which is commonly used in bacteria. | 17 | |
| BBa_K2596010 | RBS+Spacer from PpsbA2 (Synechocystis sp. 6803) | 17 | |
| BBa_K2598999 | 111 | 4089 | |
| BBa_K2619012 | internal ribosome entry site/IRES from Cloning vector pPKm-244 | 578 | |
| BBa_K2621038 | CAT-Seq Ribosome Binding Site (RBS) | 6 | |
| BBa_K2633004 | RBS + scar | 30 | |
| BBa_K2656008 | Very Weak Ribosome Binding Site J61100 | 24 | |
| BBa_K2656009 | Strong Ribosome Binding Site B0030 | 25 | |
| BBa_K2656010 | Weak Ribosome Binding Site B0032 | 27 | |
| BBa_K2656011 | Medium Ribosome Binding Site B0034 | 22 | |
| BBa_K2656012 | Very Low Ribosome Binding Site J61101 | 22 | |
| BBa_K2675010 | Synthetic RBS designed for AimR of phage phi3T | 44 | |
| BBa_K2675011 | Synthetic RBS designed for AimP of Phi3T | 38 | |
| BBa_K2675012 | Synthetic RBS designed for OmpA-SAIRGA | 41 | |
| BBa_K2675013 | Synthetic RBS designed for PelB-SAIRGA | 38 | |
| BBa_K2675014 | Synthetic RBS designed for Tat-SAIRGA | 35 | |
| BBa_K2675015 | Synthetic RBS designed for sfGFP | 45 | |
| BBa_K2675016 | Synthetic RBS designed for sfGFP | 30 | |
| BBa_K2675017 | Synthetic RBS designed for sfGFP | 30 | |
| BBa_K2680529 | BCD12 | 84 | |
| BBa_K2680530 | B0033m | 20 | |
| BBa_K2680531 | B0032m | 22 | |
| BBa_K2680561 | BCD8 | 84 | |
| BBa_K2685025 | RBS | 12 | |
| BBa_K2686007 | T7 RBS | 36 | |
| BBa_K2715009 | C.acetobutylicum thiolase gene RBS | 27 | |
| BBa_K2715020 | C.sporogenes ferredoxin gene RBS | 36 | |
| BBa_K2715022 | C.difficile Toxin A promoter RBS | 29 | |
| BBa_K2715024 | C.difficile Toxin B promoter RBS | 30 | |
| BBa_K2717029 | RBS2000 reverse | 28 | |
| BBa_K2717030 | RBS2000 | 28 | |
| BBa_K2753036 | RBSsp1 | 20 | |
| BBa_K2753037 | RBSsp2 | 20 | |
| BBa_K2753053 | RBStrGPPS | 56 | |
| BBa_K2753054 | Charles RBS | 14 | |
| BBa_K2753070 | RBSobGES | 24 | |
| BBa_K2760006 | RBS for Lactobacillus rhamnosus GG | 45 | |
| BBa_K2761006 | RBS for Cas1-Cas2 | 12 | |
| BBa_K2765024 | 1000bp Homologous arm+ura(△yno1) | 2804 | |
| BBa_K2779906 | RBS from pBAD | 33 | |
| BBa_K2782000 | Ribosome binding site preceding the repressor protein sequence of the fatty acid operon | 29 | |
| BBa_K2782003 | Ribosome binding site before the red fluorescent protein sequence | 15 | |
| BBa_K2782009 | Glyoxylate operon ribosome binding site | 15 | |
| BBa_K2787035 | orthogonal ribosome binding site A2 | 17 | |
| BBa_K2787036 | orthogonal ribosome binding site B8 | 18 | |
| BBa_K2787037 | orthogonal ribosome binding site C9 | 18 | |
| BBa_K2796071 | Its a RBS | ||
| BBa_K2797000 | RBS for streptomycin resistance | 12 | |
| BBa_K2800016 | RBS2 | 15 | |
| BBa_K2800019 | RBS1 | 12 | |
| BBa_K2800031 | RBS strength is 10 | 24 | |
| BBa_K2800032 | RBS strength is 1000 | 27 | |
| BBa_K2800033 | RBS strength is 10000 | 26 | |
| BBa_K2800034 | RBS strength is maximum | 20 | |
| BBa_K2809001 | Kozak sequence (Pichia pastoris) | 7 | |
| BBa_K2814014 | complement of BBa_B0034 | 15 | |
| BBa_K2842671 | RBS | 24 | |
| BBa_K2842684 | RBS | 22 | |
| BBa_K2842698 | RBS afafafafowoeodek | 27 | |
| BBa_K2842704 | RBS | 18 | |
| BBa_K285201 | mrgA rbs | 27 | |
| BBa_K2857013 | P1-GFP RBS (Salis lab) | 25 | |
| BBa_K2857014 | P2-GFP RBS (Salis lab) | 36 | |
| BBa_K2857015 | P3-GFP RBS (Salis lab) | 35 | |
| BBa_K2857016 | P4-GFP RBS (Salis lab) | 37 | |
| BBa_K2857017 | P5-GFP RBS (Salis lab) | 26 | |
| BBa_K2857018 | P6-GFP RBS (Salis lab) | 27 | |
| BBa_K2857019 | P70-GFP RBS (Salis lab) | 20 | |
| BBa_K2857020 | P71-GFP RBS (Salis lab) | 24 | |
| BBa_K2857021 | P9-GFP RBS (Salis lab) | 29 | |
| BBa_K2857022 | P8-GFP RBS (Salis lab) | 23 | |
| BBa_K2888011 | Test RBS | 600 | |
| BBa_K2906017 | RBS from T7 bacteriophage | 8 | |
| BBa_K2918013 | BBa_B0032 RBS and SarJ | 98 | |
| BBa_K2918014 | Universal RBS | 10 | |
| BBa_K2918035 | BBa_B0032 RBS and RiboJ | 94 | |
| BBa_K2918036 | SarJ and Universal RBS | 89 | |
| BBa_K2918038 | RiboJ and Universal RBS | 85 | |
| BBa_K2924009 | RBS* | 24 | |
| BBa_K2924053 | RBS from pET22b | 44 | |
| BBa_K2927001 | Ribosome binding site | 23 | |
| BBa_K2933024 | RBS | 7 | |
| BBa_K2933039 | RBS a | 6 | |
| BBa_K2934007 | 5' UTR derived from pBE-S for Bacillus subtilis | 40 | |
| BBa_K2935004 | nnn | 44 | |
| BBa_K294120 | nasa | ||
| BBa_K2942701 | a kind of riboswitch developed from the hammerhead ribozyme from the satellite RNA of tobacco ringsp | 125 | |
| BBa_K2943900 | Improved RBS | 21 | |
| BBa_K2943903 | an improved rbs | 21 | |
| BBa_K2943905 | an improved rbs | 19 | |
| BBa_K2963006 | RBS - recognized by Corynebacterium glutamicum | 9 | |
| BBa_K2963010 | RBS18 - a designed SD sequence for racE | 36 | |
| BBa_K2963011 | RBS20 - a designed SD sequence for racE | 35 | |
| BBa_K2963025 | RBS180 - a designed SD sequence for racE | 36 | |
| BBa_K2963026 | RBS200 - a designed SD sequence for racE | 33 | |
| BBa_K2963027 | RBS2000 - a designed SD sequence for racE | 33 | |
| BBa_K2963028 | RBS18000 - a designed SD sequence for racE | 31 | |
| BBa_K2963034 | RBS-H - recognized by Corynebacterium glutamicum | 27 | |
| BBa_K2963035 | RBS-L - recognized by Corynebacterium glutamicum | 25 | |
| BBa_K2967001 | strong rbs site | 28 | |
| BBa_K2969990 | RBS=3000 | 24 | |
| BBa_K2969994 | RBS-4 | 14 | |
| BBa_K2969997 | RBS-3 | 14 | |
| BBa_K2969998 | RBS-2 | 32 | |
| BBa_K2969999 | RBS-1 | 33 | |
| BBa_K2971011 | Ribosomal binding site | 42 | |
| BBa_K2974701 | trial 1 | 9 | |
| BBa_K2978102 | Strong RBS | 18 | |
| BBa_K2978207 | Lactobacillus spp Ribosomal Binding Site | 17 | |
| BBa_K2978306 | Ribosomal Binding Site Reverse Complement | 17 | |
| BBa_K2985008 | RBS->a designed SD sequence for sacB | 18 | |
| BBa_K2992011 | RBS region for botR gene from C. botulinum | 20 | |
| BBa_K2992031 | RBS region for the ctfA gene from C. acetobutylicum | 20 | |
| BBa_K2992032 | RBS region for the ctfA gene from C. botulinum | 20 | |
| BBa_K2992033 | RBS region for the adc gene from C. acetobutylicum. | 20 | |
| BBa_K3003044 | RBS | 12 | |
| BBa_K3027005 | RBS in pBAD24-MoClo (Golden Gate compatible) | 12 | |
| BBa_K3033002 | Ribosome binding site (pET11a) | 23 | |
| BBa_K3033003 | Ribosome binding site (pBAD24) | 31 | |
| BBa_K3059412 | MSMEG_2389 5' UTR | 39 | |
| BBa_K3059413 | MSMEG_3050 5' UTR | 74 | |
| BBa_K3059601 | RBSII | 9 | |
| BBa_K3059608 | RBSG | 19 | |
| BBa_K3059615 | RBSH | 17 | |
| BBa_K3059620 | pre-csgE RBS | 38 | |
| BBa_K3076801 | RBS derived from pET151 expression vector | 23 | |
| BBa_K3078008 | RBS in front of hpaA | 9 | |
| BBa_K3078011 | RBS from pVE5523 | 95 | |
| BBa_K3083005 | RBS for pBAD promoter | 41 | |
| BBa_K3083008 | Ptac RBS | 21 | |
| BBa_K3086004 | Ribosome binding site | 19 | |
| BBa_K3086006 | Ribosome binding site | 36 | |
| BBa_K3096000 | RBS | 15 | |
| BBa_K3098004 | Rbs of Gluconacetobacter xylinus | 13 | |
| BBa_K3105111 | RBS with ribosome standby site | 30 | |
| BBa_K3113003 | kozak | 9 | |
| BBa_K3120010 | IRES | 576 | |
| BBa_K3127007 | RBS for CcaS | 13 | |
| BBa_K3135002 | (Elowitz 1999) -- defines RBS efficiency | 12 | |
| BBa_K3135003 | x0 | 59 | |
| BBa_K3135004 | y0 | 38 | |
| BBa_K3135011 | y1 | 37 | |
| BBa_K3135012 | y2 | 36 | |
| BBa_K3135013 | y3 | 35 | |
| BBa_K3135014 | y4 | 39 | |
| BBa_K3135015 | y5 | 40 | |
| BBa_K3135016 | y6 | 41 | |
| BBa_K3135017 | y7 | 42 | |
| BBa_K3135018 | y8 | 43 | |
| BBa_K3135019 | y9 | 38 | |
| BBa_K3135020 | y10 | 38 | |
| BBa_K3135021 | y11 | 38 | |
| BBa_K3135022 | y12 | 38 | |
| BBa_K3135023 | y13 | 38 | |
| BBa_K3135024 | y14 | 38 | |
| BBa_K3135025 | y15 | 39 | |
| BBa_K3135026 | y16 | 39 | |
| BBa_K3135027 | y17 | 39 | |
| BBa_K3135028 | y18 | 39 | |
| BBa_K3135029 | y19 | 39 | |
| BBa_K3136003 | B0034-typeIIS | 12 | |
| BBa_K3136010 | B0034-RBS-A | 18 | |
| BBa_K3136011 | B0034-RBS-B | 17 | |
| BBa_K3136012 | B0034-RBS-C | 26 | |
| BBa_K3136013 | B0034-RBS-D | 25 | |
| BBa_K3137001 | Improved RBS 4 | 11 | |
| BBa_K3143683 | RBS-1 | 18 | |
| BBa_K3143685 | RBS-2 | 15 | |
| BBa_K3143687 | RBS-3 | 31 | |
| BBa_K3158003 | B0034 | 12 | |
| BBa_K3158025 | B0034-RBS-A | 18 | |
| BBa_K3158027 | B0034-RBS-B | 17 | |
| BBa_K3158029 | B0034-RBS-C | 26 | |
| BBa_K3158031 | B0034-RBS-D | 25 | |
| BBa_K3165004 | sRBS (250) | 34 | |
| BBa_K3165005 | sRBS (30000) | 35 | |
| BBa_K3183020 | Operon Spacer for E. coli | 32 | |
| BBa_K3183026 | Spacer between AgrA and AgrC | 65 | |
| BBa_K3189007 | RBS | 36 | |
| BBa_K3192000 | Custom RBS Optimized for BBa_K3192020 | 25 | |
| BBa_K3192001 | Custom RBS Optimzed for BBa_K3192021 | 25 | |
| BBa_K3192002 | Custom RBS Optimzed for BBa_K3192022 | 25 | |
| BBa_K3192004 | Custom RBS Optimzed for BBa_K3192015 | 25 | |
| BBa_K3192005 | Custom RBS Optimzed for BBa_K3192016 | 25 | |
| BBa_K3192006 | Custom RBS Optimzed for BBa_K3192017 | 25 | |
| BBa_K3192007 | Custom RBS Optimzed for BBa_K3192019 | 25 | |
| BBa_K3192008 | Custom RBS Optimzed for BBa_K2259018 | 54 | |
| BBa_K3192009 | Custom RBS Optimzed for BBa_K2259019 | 32 | |
| BBa_K3192030 | Custom RBS Optimzed for BBa_K3192018 | 25 | |
| BBa_K3196001 | Kozak | 11 | |
| BBa_K3197002 | oxidase->RBS | 20 | |
| BBa_K3202021 | RBS1 | 45 | |
| BBa_K3202026 | RBS2 | 38 | |
| BBa_K3202028 | B0034 | 12 | |
| BBa_K3206013 | pET RBS | 13 | |
| BBa_K3215000 | phnE RBS | 54 | |
| BBa_K3222002 | RBS for PURE system | 14 | |
| BBa_K3228054 | pMC_0_3_12_riboJ_B0034 | 97 | |
| BBa_K3228055 | pMC_0_3_13_riboJ_B0034_noScar | 91 | |
| BBa_K3228056 | pMC_0_3_14_SarJ_B0034 | 101 | |
| BBa_K3228057 | pMC_0_3_15_PlmJ_B0034 | 104 | |
| BBa_K3229294 | BBa_0030 RBS reverse complement | 15 | |
| BBa_K3247012 | Type IIS compatible BBa_B0034 with RFC10 scar context | 23 | |
| BBa_K3252025 | Medium Ribosome Binding Site BBa_B0034 | 12 | |
| BBa_K3252026 | Medium Ribosome Binding Site BBa_B0032 | 13 | |
| BBa_K3252027 | Very Weak Ribosome Binding Site BBa_B0033 | 11 | |
| BBa_K3254019 | A RBS used in BBa_K3254011 | 25 | |
| BBa_K3254020 | A RBS used in BBa_K3254012 | 27 | |
| BBa_K3254021 | A RBS used in BBa_K3254013 | 34 | |
| BBa_K3254028 | A RBS used in BBa_K3254026 | 21 | |
| BBa_K3254029 | A RBS used in BBa_K3254027 | 40 | |
| BBa_K3257010 | Bacterial RBS | 12 | |
| BBa_K3257011 | T7 RBS | 23 | |
| BBa_K3265022 | RBS | 63 | |
| BBa_K3286013 | RBS_11 | 14 | |
| BBa_K3286014 | RBS_C7 | 14 | |
| BBa_K3286015 | RBS_B10 | 14 | |
| BBa_K3286016 | RBS_D2 | 15 | |
| BBa_K3286017 | RBS_B6 | 14 | |
| BBa_K3286018 | RBS_A12 | 14 | |
| BBa_K3286019 | RBS_A9 | 14 | |
| BBa_K3288005 | Cdog (strong rbs) | 95 | |
| BBa_K3288006 | NO29 (strong abs) | 42 | |
| BBa_K3288007 | strong RBS derived from pDawn | 38 | |
| BBa_K3296002 | B0034 RBS | 12 | |
| BBa_K3308042 | T7 derived Ribosome Binding Site | 16 | |
| BBa_K3308062 | T7 Translational Ribosome Binding Site | 17 | |
| BBa_K3308094 | pKLD66 (pTEV6) Ribosome Binding Site | 17 | |
| BBa_K3317015 | RBS designed for Protocatechuate decarboxylase (aroY) with sp | 29 | |
| BBa_K3317016 | RBS designed for Catechol 1,2-dioxygenase (catA) with ps | 31 | |
| BBa_K3317017 | RBS designed for Vanillate O-demethylase (LigM gene) | ||
| BBa_K3317018 | RBS designed for Vanillate O-demethylase (LigM gene) with sp | 33 | |
| BBa_K3317019 | RBS designed for pduU mod | 36 | |
| BBa_K3317021 | RBS designed for VanR repressor | 35 | |
| BBa_K3317022 | RBS designed for pCaU repressor | 38 | |
| BBa_K3317055 | RBS designed to maximize NucE expression in E.coli K12 | 32 | |
| BBa_K3317056 | RBS designed to maximize NucD expression in E.coli K12 | 32 | |
| BBa_K3317057 | RBS designed to maximize NucA expression in E.coli K12 | 35 | |
| BBa_K3317058 | RBS designed for φX174E overexpression in E.coli K12 | 35 | |
| BBa_K3317067 | 37 į C inducible RNA thermometer | 121 | |
| BBa_K3317068 | Synthetic RBS in pMF35 | 24 | |
| BBa_K3317072 | MF002 | 24 | |
| BBa_K3320012 | RBS (J. Anderson, 2005) | 13 | |
| BBa_K3331026 | araBAD RBS | 7 | |
| BBa_K3332036 | RBS1 | 9 | |
| BBa_K3338004 | Internal ribosome entry site (IRES) for use in eukaryotic cells | 574 | |
| BBa_K3351001 | A ribosome binding site from pet28a plasmid. | 23 | |
| BBa_K3352002 | Extended RBS | 23 | |
| BBa_K3356000 | RBS region for bdhA | 20 | |
| BBa_K3360006 | synthetic rbs | 27 | |
| BBa_K3365002 | RBS | 20 | |
| BBa_K3386005 | RBS associated with pMphR | 12 | |
| BBa_K3395003 | RBS | 23 | |
| BBa_K3395012 | RBS2 | 28 | |
| BBa_K3425031 | RBS for Type IIS iGEM Standard Assembly | 18 | |
| BBa_K3425032 | RBS for Type IIS iGEM Standard Assembly | 17 | |
| BBa_K3425033 | RBS for Type IIS iGEM Standard Assembly | 20 | |
| BBa_K3425048 | Juniper GFP for Type IIS iGEM standard assembly | 705 | |
| BBa_K3453005 | Synthetic RBS stem-loop designed for standard toehold switches | 25 | |
| BBa_K3457007 | RBS0034 | 12 | |
| BBa_K3463003 | RBS eGFP and RhlR (RhlR construction) | 25 | |
| BBa_K3463009 | RBS eGFP | 25 | |
| BBa_K3463010 | RBS LacI | 25 | |
| BBa_K3463012 | Ptac RBS GFP low | 25 | |
| BBa_K3463013 | Ptac RBS GFP high | 25 | |
| BBa_K3463014 | Ptac RBS GFP low | 25 | |
| BBa_K3463015 | Ptac RBS E7 50% | 25 | |
| BBa_K3463016 | Ptac RBS E7 25% | 25 | |
| BBa_K3463021 | RBS mCherry High | 26 | |
| BBa_K3463022 | RBS mCherry low | 26 | |
| BBa_K3498014 | Evolved RBS for VanRAM | 29 | |
| BBa_K3498015 | RBS for Rsn-2 | 34 | |
| BBa_K3498016 | RBS for Rsn-3 | 25 | |
| BBa_K3498017 | RBS for Rsn-4 | 25 | |
| BBa_K3498018 | RBS for Rsn-5 | 25 | |
| BBa_K3498019 | RBS for PhrC | 25 | |
| BBa_K3498020 | RBS for sfp | 25 | |
| BBa_K3498021 | RBS for Spo0E | 25 | |
| BBa_K3498022 | RBS for AbrB | 25 | |
| BBa_K3498023 | RBS for SinI | 25 | |
| BBa_K3498024 | RBS for AbbA | 25 | |
| BBa_K3520008 | Flavobacteria RBS | 5 | |
| BBa_K3552030 | GGAGG | 5 | |
| BBa_K3560003 | DrRBS | 5 | |
| BBa_K356105 | Desinged RBS | 14 | |
| BBa_K356106 | Desinged RBS | 16 | |
| BBa_K3579001 | RBS_sodA | 64 | |
| BBa_K3579003 | EpiFlex_SaTIR | 74 | |
| BBa_K3579004 | EpiFlex_gsiB | 63 | |
| BBa_K3584007 | RBS in tyrP | 29 | |
| BBa_K3585000 | the RBS in pMTL83151 | 14 | |
| BBa_K3587002 | RBS for Ada protein | ||
| BBa_K3587003 | RBS for Ada protein | ||
| BBa_K3587004 | RBS for Ada protein | 15 | |
| BBa_K3590001 | RBS R0 | 24 | |
| BBa_K3590002 | RBS R1 | 21 | |
| BBa_K3590003 | RBS R2 | 20 | |
| BBa_K3594100 | an interesting gene | 49 | |
| BBa_K3596002 | hofB RBS | 20 | |
| BBa_K3596004 | hofM RBS | 44 | |
| BBa_K3596006 | ppdA RBS | 18 | |
| BBa_K3596008 | ppdA RBS mut1 | 18 | |
| BBa_K3596009 | ppdA RBS mut2 | 18 | |
| BBa_K3596010 | hofB RBS mut1 | 20 | |
| BBa_K3596011 | hofB RBS mut2 | 20 | |
| BBa_K3596012 | hofM RBS mut | 44 | |
| BBa_K3598014 | RBS | 20 | |
| BBa_K3598023 | RBS0 | 20 | |
| BBa_K3598024 | RBS1 | 20 | |
| BBa_K3598025 | RBS3 | 20 | |
| BBa_K3598026 | RBS5 | 20 | |
| BBa_K3598027 | RBS7 | 20 | |
| BBa_K3598028 | RBS9 | 20 | |
| BBa_K3598029 | RBS11 | 20 | |
| BBa_K3598030 | RBS13 | 20 | |
| BBa_K3598031 | RBS15 | 20 | |
| BBa_K3598032 | RBS15 | 20 | |
| BBa_K3598033 | RBS19 | 20 | |
| BBa_K3598034 | RBS21 | 20 | |
| BBa_K3598035 | RBS23 | 20 | |
| BBa_K3598036 | RBS25 | 20 | |
| BBa_K3598037 | RBS27 | 20 | |
| BBa_K3598038 | RBS29 | 20 | |
| BBa_K3599000 | Ribosome Binding Site | 23 | |
| BBa_K3599013 | RBS 2 | 12 | |
| BBa_K360031 | Strong RBS with spacer. | 11 | |
| BBa_K3601004 | Strong RBS | 23 | |
| BBa_K3612029 | RBS of Pasp promoter | 6 | |
| BBa_K3612030 | RBS of pSHAB | 4 | |
| BBa_K3612031 | RBS of pMAV promoter | 72 | |
| BBa_K3628013 | RBS1 between HpaB and HpaC | 17 | |
| BBa_K3628014 | RBS2 control HpaB | 45 | |
| BBa_K3628015 | RBS3 between HpaB SMS and HpaC SMS | 34 | |
| BBa_K3630002 | A ribosome binding site, or ribosomal binding site (RBS) | 6 | |
| BBa_K3665006 | RBS4 ,Regulating the expression of atLCD | 25 | |
| BBa_K3665007 | Regulating the expression of atLCD | 25 | |
| BBa_K3665008 | Regulating the expression of atLCD | 25 | |
| BBa_K3712012 | pR/pL RBS | 11 | |
| BBa_K3715082 | RBS | 7 | |
| BBa_K3716001 | RBS(pet28a) | 23 | |
| BBa_K3726006 | RBS_PhaB^173S | 25 | |
| BBa_K3726007 | RBS_ccr | 26 | |
| BBa_K3726008 | RBS_PhaJ | 38 | |
| BBa_K3726009 | RBS_PduP_MediumExpression | 25 | |
| BBa_K3726010 | RBS_Slr1192_MediumExpression | 25 | |
| BBa_K3726020 | RBS_pKpA | 33 | |
| BBa_K3726022 | RBS_mdh | ||
| BBa_K3726024 | RBS_garK | 29 | |
| BBa_K3726026 | RBS_garR | 25 | |
| BBa_K3726030 | RBS_gcl | 25 | |
| BBa_K3726032 | RBS_mtkA | 31 | |
| BBa_K3726034 | RBS_mtkB | 25 | |
| BBa_K3726036 | RBS_mcl | 25 | |
| BBa_K3726082 | RBS BCD2(Mod) | ||
| BBa_K3726086 | RBS BCD2(Mod) | 93 | |
| BBa_K3726088 | RBS_NphT7 | 33 | |
| BBa_K3726089 | RBS_Slr1192_HighExpression | 25 | |
| BBa_K3726090 | RBS_PduP | 38 | |
| BBa_K3726091 | RBS_ppc | 33 | |
| BBa_K3726092 | RBS_PtaBs | 55 | |
| BBa_K3726093 | RBS* | 10 | |
| BBa_K3740008 | RBS009 | 12 | |
| BBa_K3740010 | RBS010 | 12 | |
| BBa_K3740018 | RBS004 | 12 | |
| BBa_K3740025 | RBS150 | 28 | |
| BBa_K3740026 | RBS300 | 27 | |
| BBa_K3740027 | RBS II | 10 | |
| BBa_K3740028 | RBS1000 | 27 | |
| BBa_K3740039 | RBS400 | 24 | |
| BBa_K3745004 | RBS on pSC101 | 12 | |
| BBa_K3745005 | EC6015/Bujard RBS for biosensor | 57 | |
| BBa_K3758070 | atpB 5-UTR (Nicotiana tabacum) | 255 | |
| BBa_K3758071 | atpB 5-UTR (+10) (Nicotiana tabacum) | 265 | |
| BBa_K3758072 | atpB 5-UTR (-90 to +10 processed) (Nicotiana tabacum) | 100 | |
| BBa_K3758073 | atpB 5-UTR (TCR processed -90) (Nicotiana tabacum) | 132 | |
| BBa_K3758074 | atpH 5-UTR (-45 processed) (Nicotiana tabacum) | 45 | |
| BBa_K3758075 | psbN 5-UTR (Nicotiana tabacum) | 47 | |
| BBa_K3758076 | psbN 5-UTR (processed) (Nicotiana tabacum) | 39 | |
| BBa_K3758077 | psbB 5-UTR (Nicotiana tabacum) | 175 | |
| BBa_K3758078 | ndhD 5-UTR (Nicotiana tabacum) | 118 | |
| BBa_K3758079 | rps2 mAAGA 5-UTR (Nicotiana tabacum) | 42 | |
| BBa_K3758080 | rps2 mCCUC 5-UTR (Nicotiana tabacum) | 42 | |
| BBa_K3758081 | rps2 mAAGA 5-UTR (-20 to -10 deletion) (Nicotiana tabacum) | 32 | |
| BBa_K3758082 | rps2 mCCUC 5-UTR (-20 to -10 deletion) (Nicotiana tabacum) | 32 | |
| BBa_K3758083 | psbH 5-UTR (Startcodon + 1AA) (Nicotiana tabacum) | 58 | |
| BBa_K3758084 | psbA 5-UTR (SNM) (Nicotiana tabacum) | 88 | |
| BBa_K3758085 | psbA 5-UTR (SEM) (Nicotiana tabacum) | 88 | |
| BBa_K3758086 | psbA 5-UTR (deltaSL) (Nicotiana tabacum) | 53 | |
| BBa_K3758087 | psbA 5-UTR (delta5-SL) (Nicotiana tabacum) | 48 | |
| BBa_K3758088 | psbA-rbcL 5-UTR (R-SLWT) (Nicotiana tabacum) | 108 | |
| BBa_K3758089 | rps14 5-UTR (Nicotiana tabacum) | 122 | |
| BBa_K3758090 | rbcL 5-UTR (Nicotiana tabacum) | 182 | |
| BBa_K3758091 | rbcL 5-UTR (processed) (Nicotiana tabacum) | 58 | |
| BBa_K3758092 | rbcL 5-UTR (+14AA) (Nicotiana tabacum) | 100 | |
| BBa_K3758093 | rrn16 5-UTR (Nicotiana tabacum) | 114 | |
| BBa_K3758094 | rrn16 5-UTR (+37) (Nicotiana tabacum) | 37 | |
| BBa_K3758095 | psbA 5-UTR (Nicotiana tabacum) | 88 | |
| BBa_K3758096 | Synthetic 5-UTR (Nicotiana tabacum) | 56 | |
| BBa_K3758097 | psbC 5-UTR (Nicotiana tabacum) | 230 | |
| BBa_K3758098 | accD 5-UTR (Nicotiana tabacum) | 180 | |
| BBa_K3758099 | clpP 5-UTR (Nicotiana tabacum) | 48 | |
| BBa_K3758100 | rbcL 5-UTR (Spinacia oleracea) | 162 | |
| BBa_K3758101 | ndhC 5-UTR (Spinacia oleracea) | 270 | |
| BBa_K3758102 | ndhB 5-UTR (Spinacia oleracea) | 214 | |
| BBa_K3758103 | atpB 5-UTR (Spinacia oleracea) | 453 | |
| BBa_K3758104 | psaC 5-UTR (Oryza sativa) | 264 | |
| BBa_K3758105 | psbA 5-UTR (Oryza sativa) | 78 | |
| BBa_K3758106 | psbB 5-UTR (Oryza sativa) | 197 | |
| BBa_K3758107 | rpl23 5-UTR (Oryza sativa) | 73 | |
| BBa_K3758108 | rpl32 5-UTR (Oryza sativa) | 198 | |
| BBa_K3758109 | rps12 5-UTR (Oryza sativa) | 397 | |
| BBa_K3758110 | rps19 5-UTR (Oryza sativa) | 107 | |
| BBa_K3758111 | T7 gene10 5-UTR | 63 | |
| BBa_K3758113 | rpl33 5'UTR Triticum aestivum | 277 | |
| BBa_K3758114 | rps4 5-UTR (Triticum aestivum) | 128 | |
| BBa_K3758115 | rps12 5-UTR (Triticum aestivum) | 222 | |
| BBa_K3758116 | rps19 5-UTR (Triticum aestivum) | 130 | |
| BBa_K3758117 | rbcL 5-UTR (full) Triticum aestivum | 317 | |
| BBa_K3758118 | psbA 5-UTR Triticum aestivum | 81 | |
| BBa_K3758119 | atpB 5-UTR Triticum aestivum | 297 | |
| BBa_K3758120 | psaC 5-UTR Tritcum aestivum | 377 | |
| BBa_K3758121 | atpB 5-UTR (Quercus robur) | 506 | |
| BBa_K3758122 | psbA 5-UTR (Quercus robur) | 92 | |
| BBa_K3758123 | rbcL 5-UTR (Quercus robur) | 140 | |
| BBa_K3758124 | Synthetic 5-UTR | 115 | |
| BBa_K3758125 | Synthetic 5-UTR 2 | 125 | |
| BBa_K3758126 | Synthetic 5-UTR 3 | 86 | |
| BBa_K3758127 | Synthetic 5-UTR 4 | 51 | |
| BBa_K376001 | Super RBS | 24 | |
| BBa_K3763000 | RBS improved on the basis of BBa_B0030 | 15 | |
| BBa_K3763046 | RBS(EBW25113/RBS_fumB) | 52 | |
| BBa_K3766031 | RBS for TetR in reverse orientation | 18 | |
| BBa_K3766032 | RBS from pET26b with XbaI and SalI | 29 | |
| BBa_K3766033 | Synthetic RBS designed for mCherry (reverse orientation) | 24 | |
| BBa_K3766034 | Synthetic RBS designed for sfGFP | 27 | |
| BBa_K3766035 | Synthetic RBS designed for LacI | 29 | |
| BBa_K3766190 | LacI in the Evolution.T7 mutational region | 1694 | |
| BBa_K3769011 | T7RBS | 23 | |
| BBa_K3773007 | clpB RBS native to E. coli | 9 | |
| BBa_K3773008 | RBS-containing region provided by Ceroni et al., 2015 for TL sensor | 32 | |
| BBa_K3773015 | RBS-containing region following P70a | 35 | |
| BBa_K3777030 | 1 | 28 | |
| BBa_K3778013 | RBS (Ribosome-binding site ) | 23 | |
| BBa_K3782013 | crRNA leader sequence | 133 | |
| BBa_K3782018 | cspA 5′-UTR | 134 | |
| BBa_K3790084 | RBS for Gram-positive species (Schofield, 2003) | 17 | |
| BBa_K3793001 | Ribosomal Binding site for E. coli | 12 | |
| BBa_K3796001 | Corynebacterium glutamicum strong RBS | 9 | |
| BBa_K3796005 | Corynebacterium glutamicum weak RBS | 10 | |
| BBa_K3796212 | A strong strength RBS | 9 | |
| BBa_K3798018 | RBS for B. subtilis | 5 | |
| BBa_K3799051 | DEMO 2 | 134 | |
| BBa_K3812011 | RBS | 64 | |
| BBa_K3814011 | PilE + RBS, ACIAD3314 | 447 | |
| BBa_K3814047 | RBS for TcR gene | 9 | |
| BBa_K3820010 | Xanthomonas Oryzae RBS | 20 | |
| BBa_K3822000 | Toehold Switch | 84 | |
| BBa_K3831005 | Synthetic RBS_a | 22 | |
| BBa_K3831006 | Synthetic RBS_b | 33 | |
| BBa_K3831007 | Native RBS R2 | 21 | |
| BBa_K3831008 | RBS R6 | 22 | |
| BBa_K3831009 | Synthetic RBS_c | 27 | |
| BBa_K3831014 | Native RBS R1 | 21 | |
| BBa_K3831018 | Native RBS R0 | 24 | |
| BBa_K3831020 | Artificial RBS R3 | 22 | |
| BBa_K3831047 | Synthetic RBS_c - reversed | 27 | |
| BBa_K3831049 | RBS R6 - reversed | 22 | |
| BBa_K3831051 | RBS R2 - reversed | 21 | |
| BBa_K3831053 | Synthetic RBS_b - reversed | 33 | |
| BBa_K3831055 | Synthetic RBS_a - reversed | 22 | |
| BBa_K3832003 | medium RBS | 10 | |
| BBa_K3832004 | medium RBS | 14 | |
| BBa_K3832005 | weak RBS | 11 | |
| BBa_K3842013 | RBS | 25 | |
| BBa_K3846102 | Strong RBS (Phytobrick) | 20 | |
| BBa_K3854000 | Yeast RBS | 12 | |
| BBa_K3858008 | PYD1 RBS | ||
| BBa_K3858009 | PYD1 RBS | 40 | |
| BBa_K3861009 | sicP RBS | 46 | |
| BBa_K3861014 | fimH RBS | 14 | |
| BBa_K3875028 | RBS: aggaga | 6 | |
| BBa_K3875029 | RBS: aacaattgaaattattcctc | 21 | |
| BBa_K3885111 | UTR1 | 40 | |
| BBa_K3885112 | UTR2 | 35 | |
| BBa_K3885113 | UTR3 | 35 | |
| BBa_K3892009 | Spacer-RBS-Spacer | 45 | |
| BBa_K3892010 | G-10 leader and RBS | 36 | |
| BBa_K3893019 | Reverse of BBa_B0034 / RBS2 o RBS | 12 | |
| BBa_K3893021 | Reverse of BBa_K3893000 / ShineDalgarno for E. coli | 11 | |
| BBa_K3893022 | Reverse of BBa_B0032 / RBS3 | 13 | |
| BBa_K3895013 | 5' UTR/RBS_KerA | 59 | |
| BBa_K3898200 | P-M122 | 729 | |
| BBa_K3898201 | 34 | 3 | |
| BBa_K3898204 | 33 | 3 | |
| BBa_K390001 | 5'UTR and native RBS from psbA2 in Synechocystis sp. PCC 6803 | 49 | |
| BBa_K390002 | 5'UTR and consensus RBS from psbA2 in Synechocystis sp. PCC 6803 | 49 | |
| BBa_K3910010 | Ribosomal Binding Site 3 | 11 | |
| BBa_K3910998 | P1 Ribosome Binding Site | 22 | |
| BBa_K3914007 | Ribosome bingding site of Esa box | 12 | |
| BBa_K3929035 | RBS_pSEVA238-Intimin | 6 | |
| BBa_K3929040 | RBS_pSEVA238-Intimin | ||
| BBa_K3929041 | RBS_pSEVA228-Intimin | 4 | |
| BBa_K393002 | short RBS | 12 | |
| BBa_K3933008 | RBS optimized for TetR | 48 | |
| BBa_K3933009 | RBS optimized for Ldh | 50 | |
| BBa_K3937009 | RBS_on_pHT01 | 8 | |
| BBa_K3940012 | RBS T7 Gene 10 | 23 | |
| BBa_K3946015 | Trc-RBS (RN) | 67 | |
| BBa_K3972006 | 5-UTR with g10-L RBS | 54 | |
| BBa_K3978043 | neget-cI-RBS | ||
| BBa_K3978044 | neget-cIDN -RBS | ||
| BBa_K3985008 | E. coli consensus RBS | 6 | |
| BBa_K3985014 | E. coli consensus RBS | 9 | |
| BBa_K3985023 | RBS strong for B. subtilis | 21 | |
| BBa_K3985029 | E. coli RBS | 12 | |
| BBa_K4011990 | RBS test | 4 | |
| BBa_K4011992 | zsrbs | 12 | |
| BBa_K4011995 | zsrbs2 | 8 | |
| BBa_K4017002 | Ribosome Binding Site | 12 | |
| BBa_K4019007 | RBS (upstream of 4-coumarate-CoA ligase) | 39 | |
| BBa_K4023002 | RBS | 15 | |
| BBa_K4046050 | Kozak Sequence (Human RBS) | 6 | |
| BBa_K4054001 | T7 RBS | 23 | |
| BBa_K4056013 | Reverse RBS with 6bp spacer | 21 | |
| BBa_K4059006 | RBS for T7 promoter | ||
| BBa_K4062005 | lacZ RBS | 17 | |
| BBa_K4062007 | lacY RBS | 30 | |
| BBa_K4062018 | lacZ RBS mutation | 17 | |
| BBa_K4065001 | RBS designed for CFP protein in B. subtilis | 30 | |
| BBa_K4065003 | RBS designed for CAS9 protein in B. subtilis | 34 | |
| BBa_K4065004 | RBS designed for YokF protein in B. subtilis | 43 | |
| BBa_K4074002 | SpoVG (rbs) | 12 | |
| BBa_K4078000 | RBS Region from pTrc-trGPPS(CO)-LS | 32 | |
| BBa_K4108000 | RBS part for testing the judging form | 30 | |
| BBa_K4117666 | RNA Thermometer (U23-) | 42 | |
| BBa_K4132004 | Ribosome binding site for silicateins and urease | 6 | |
| BBa_K4133004 | RBS_tuf33 (Lactobacillus plantarum) | 76 | |
| BBa_K4133008 | RBS_Lp (Lactobacillus plantarum WCFS1) | 14 | |
| BBa_K4133011 | RBS_Lc (Lactococcus lactis) | 12 | |
| BBa_K4137015 | pET28a(+) RBS + 5' UTR | 12 | |
| BBa_K4140002 | TyrR RBS | 14 | |
| BBa_K4144001 | An intrinsic RBS in pHT01K for inserted fragments to be translated. | 10 | |
| BBa_K4144012 | An improved RBS to reduce the leakage expression of promoter Pxyl | 17 | |
| BBa_K4149222 | RBS in Escherchia | 7 | |
| BBa_K4150000 | g10.RBS | 58 | |
| BBa_K415145 | p8Acid | ||
| BBa_K4156081 | RBS spoVG | 21 | |
| BBa_K4156093 | BCD2 | 85 | |
| BBa_K4159001 | RBS (Olins and Rangwala, 1989) | 23 | |
| BBa_K4162006 | T7_RBS | 17 | |
| BBa_K4165016 | PGS-21a Ribosomal binding site | 44 | |
| BBa_K4165075 | RBS 5 | 42 | |
| BBa_K4165095 | RBS 6 | 22 | |
| BBa_K4165260 | Ubiquitin WT RBS | 24 | |
| BBa_K4165261 | RBS 1 | 43 | |
| BBa_K4165262 | RBS 2 | 61 | |
| BBa_K4165263 | RBS 3 | 41 | |
| BBa_K4165264 | RBS4 | 60 | |
| BBa_K4165265 | RBS 5 | 44 | |
| BBa_K4169023 | a theophylline-inducible riboswitch | 63 | |
| BBa_K4170018 | RBS from bacteriophage T7 gene 10 | 23 | |
| BBa_K4170053 | RBS derived from E.coli (for RhaS expression) | 24 | |
| BBa_K4171026 | RBS for tyrP | 15 | |
| BBa_K4177000 | shiyizhongqidongzi | ||
| BBa_K4190001 | Shine-Dalgarno Sequence | 23 | |
| BBa_K4192000 | Strong RBS of Pseudomonas fluorescens | 5 | |
| BBa_K4192001 | RBS of E.coli | 6 | |
| BBa_K4192002 | RBS of Pseudomonas fluorescens | 4 | |
| BBa_K4194009 | RBS | 6 | |
| BBa_K4205114 | RBS | 26 | |
| BBa_K4209015 | RBS | 9 | |
| BBa_K4209017 | RBS | 6 | |
| BBa_K4211029 | TATA | 5 | |
| BBa_K4212016 | RBS1 | 13 | |
| BBa_K4212017 | RBS2 | 13 | |
| BBa_K4212018 | RBS3 | 6 | |
| BBa_K4212019 | Optimal_RBS | 16 | |
| BBa_K4216012 | U2m (BCD11) | 84 | |
| BBa_K4216013 | U4m (BCD) -> Very Srong RBS | 34 | |
| BBa_K4216014 | U6m (BCD)->Medium Rbs (0.2-0.5) | 84 | |
| BBa_K4216015 | U8m (BCD) -> weak RBS (Weak (0.08 - 0.2)) | 84 | |
| BBa_K4216016 | U1d (NON BCD) -> Medium Weiss RBS | 22 | |
| BBa_K4216017 | U2d (NON BCD) -> Weak Weiss RBS | 20 | |
| BBa_K4216018 | U3d (NON BCD) ->Strong Weiss RBS | 21 | |
| BBa_K4229004 | RBS pCDF-Duet-1 MCS1 and MCS2 | 6 | |
| BBa_K4229005 | RBS for FRE | 25 | |
| BBa_K4229006 | RBS for TnaB | 25 | |
| BBa_K4229038 | RBS for the T proteins in the BMCs | 12 | |
| BBa_K4229039 | RBS for the H protein of the BMCs | 20 | |
| BBa_K4229058 | RBS for sfGFP | 17 | |
| BBa_K4241012 | TIR-2 translation initiation region 2 | 38 | |
| BBa_K4253010 | ribosome binding site | 12 | |
| BBa_K4256106 | ribosome binding site | 23 | |
| BBa_K4271006 | pNP RBS | 21 | |
| BBa_K4271016 | AsPhoU RBS | 21 | |
| BBa_K4274013 | Synthetic RBS1 in pJOE8999 | 8 | |
| BBa_K4274014 | Synthetic RBS2 in pJOE8999 | 8 | |
| BBa_K4274015 | Synthetic RBS1 in pHY300PLK | 18 | |
| BBa_K4274016 | Synthetic RBS1 in pHT43 | 34 | |
| BBa_K4274017 | Synthetic RBS2 in pHT43 | 23 | |
| BBa_K4274018 | Synthetic RBS3 in pHT43 | 41 | |
| BBa_K4286527 | cbh1 leader sequence | 17 | |
| BBa_K4288004 | GsiB RBS | 11 | |
| BBa_K4291004 | RBS_tnaC | ||
| BBa_K4294004 | Synthetic RBS Ath4 | 29 | |
| BBa_K4294005 | RBS Ath5 | 29 | |
| BBa_K4294010 | Synthetic RBS Ath10 | 29 | |
| BBa_K4294011 | Synthetic RBS Ath11 | 29 | |
| BBa_K4294012 | Synthetic RBS Ath12 | 28 | |
| BBa_K4294013 | Synthetic RBS Ath13 | 29 | |
| BBa_K4294101 | BCD1 | 84 | |
| BBa_K4294102 | BCD2 | 88 | |
| BBa_K4294114 | BCD14 | 88 | |
| BBa_K4294151 | RBS lac1 | 28 | |
| BBa_K4294190 | tet1 synthetic RBS | 28 | |
| BBa_K4294559 | Elowitz RBS with modified spacer region | 27 | |
| BBa_K4294596 | Elowitz RBS with modified spacer region | 26 | |
| BBa_K4294659 | Elowitz RBS with modified spacer region and an upstream Epsilon motif | 36 | |
| BBa_K4294696 | Elowitz RBS with modified spacer region and an upstream Epsilon motif | 39 | |
| BBa_K4310003 | Kozak sequence for Chlorella Vulgaris | 23 | |
| BBa_K4324200 | RBS for E. coli | 15 | |
| BBa_K4325002 | RBS070 | 12 | |
| BBa_K4325006 | RBS003422 (B0034 derivative) | 10 | |
| BBa_K4344028 | Kozak Sequence (human RBS) | 11 | |
| BBa_K4344061 | Shine-Dalgarno | 7 | |
| BBa_K4345015 | pSynSens2.5000 RBS | 35 | |
| BBa_K4348059 | ADC A RBS | 35 | |
| BBa_K4348060 | ADC D RBS | 35 | |
| BBa_K4348061 | ADC C RBS | 35 | |
| BBa_K4348062 | ismA RBS | 35 | |
| BBa_K4348063 | AKR1D1 RBS | 35 | |
| BBa_K4348064 | AKR1C4 RBS | 35 | |
| BBa_K4348120 | blah | 5 | |
| BBa_K4358015 | Ribosome binding site | 10 | |
| BBa_K4358998 | Test | ||
| BBa_K4358999 | Test | ||
| BBa_K4370007 | RBS_STREPTO | 20 | |
| BBa_K4373005 | 5'UTR including RBS for Bacillus subtilis | 75 | |
| BBa_K4382009 | RBS for Bacillus Subtilis | 15 | |
| BBa_K4387020 | Strong Ribosome Binding Site | 12 | |
| BBa_K4388011 | Strong RBS with Upstream Linker Sequence Replacing Promoter | 53 | |
| BBa_K4392000 | Ribosome binding site associated with the T7 system from the T7 bacteriophage | 57 | |
| BBa_K4401001 | RBS | 10 | |
| BBa_K4402001 | RBS | 10 | |
| BBa_K4408000 | RBS of adhE2 | ||
| BBa_K4408001 | RBS of adhE2 | 6 | |
| BBa_K4415004 | Human kozak sequence | 13 | |
| BBa_K4417008 | RBS in pCT5c | 28 | |
| BBa_K4432010 | Synthetic RBS designed for CrtW | 28 | |
| BBa_K4432011 | Synthetic RBS designed for CrtO | 27 | |
| BBa_K4432012 | Synthetic RBS designed for idi | 36 | |
| BBa_K4432013 | Synthetic RBS designed for dxs | 32 | |
| BBa_K4432014 | Synthetic RBS designed for Pseudomonas aeruginosa PhzA1-G1 operon | 27 | |
| BBa_K4432015 | Synthetic RBS designed for Pseudomonas aeruginosa PhzA2-G2 operon | 25 | |
| BBa_K4435015 | TU_RBS | 20 | |
| BBa_K4437500 | DeadRBS (non-coding) | 15 | |
| BBa_K4450008 | Ribosomal binding site | 19 | |
| BBa_K4458999 | The Alverno_Ca team characterized BBa_K2066035, which contains BBa-B0034, as well as a series of oth | 12 | |
| BBa_K4462010 | RBS.15 | 12 | |
| BBa_K4472997 | RBS 1 | 8 | |
| BBa_K4477006 | RBS for expression of human/mouse anti-apoB(MDA) antibody light chain | 27 | |
| BBa_K4477007 | RBS for expression of human/mouse anti-apoB(MDA) antibody heavy chain | 26 | |
| BBa_K4477008 | RBS for scFv expression (IK17) | 30 | |
| BBa_K4477009 | RBS for scFv expression (McPC603) | 31 | |
| BBa_K4477010 | RBS for camelid anti-mouse VHH expression | 39 | |
| BBa_K4488015 | RBS | 20 | |
| BBa_K4514043 | RBS(KanR) | 10 | |
| BBa_K4575016 | efficient ribosome binding site from bacteriophage T7 gene 10 | 23 | |
| BBa_K4578011 | Ribosome binding site for B. burgdorferi specific surface proteins | 6 | |
| BBa_K4578012 | Ribosome binding site for B. burgdorferi specific salivary protein | 6 | |
| BBa_K4587104 | RBS from pRSF | 6 | |
| BBa_K4587113 | RBS (GFP) | 6 | |
| BBa_K4587118 | vicA RBS | 6 | |
| BBa_K4591006 | Reversed RBS_BBa_B0034 | 12 | |
| BBa_K4592003 | Synthetic RBS for Microcystis-optimized eGFP Expression | 62 | |
| BBa_K4595029 | RBS-lldp | 22 | |
| BBa_K4595030 | RBS-ladhA | 22 | |
| BBa_K4595031 | RBS-dsup | 22 | |
| BBa_K4595032 | RBS-omcs | 22 | |
| BBa_K4595033 | RBS-gshA | 22 | |
| BBa_K4595034 | RBS-gshB | 22 | |
| BBa_K4595035 | RBS-ycel | 12 | |
| BBa_K4595036 | RBS-pncB | 12 | |
| BBa_K4595037 | RBS-nadE | 12 | |
| BBa_K4595038 | RBS-nadD | 12 | |
| BBa_K4595039 | RBS-nadM | 12 | |
| BBa_K4595040 | RBS-dsup | 12 | |
| BBa_K4595041 | RBS-sodA | 12 | |
| BBa_K4595042 | RBS-oprf | 14 | |
| BBa_K4601100 | Synthetic RBS designed for CatCh | 27 | |
| BBa_K4601101 | Synthetic RBS designed for ChrimsonR | 27 | |
| BBa_K4601102 | Synthetic RBS designed for Jaws | 25 | |
| BBa_K4601103 | Synthetic RBS designed for NpHR | 25 | |
| BBa_K4601105 | Synthetic RBS designed for NpSR | 25 | |
| BBa_K4601106 | Synthetic RBS designed for NpHTRII-EnvZ | 27 | |
| BBa_K4601131 | Synthetic RBS designed for LacZalpha | 28 | |
| BBa_K4601132 | Synthetic RBS designed for LacZalpha | 28 | |
| BBa_K4601133 | Synthetic RBS designed for LacZalpha | 28 | |
| BBa_K4601134 | Synthetic RBS designed for LacZalpha | 28 | |
| BBa_K4601141 | Synthetic RBS designed for AmpR | 30 | |
| BBa_K4601142 | Synthetic RBS designed for AmpR | 30 | |
| BBa_K4601143 | Synthetic RBS designed for AmpR | 30 | |
| BBa_K4601144 | Synthetic RBS designed for AmpR | 30 | |
| BBa_K4601151 | RBS for CmR | 29 | |
| BBa_K4604004 | modified T7 RBS | 14 | |
| BBa_K4604030 | RBS for piG_01a | 12 | |
| BBa_K4604038 | MazE RBS | 25 | |
| BBa_K4604044 | Riboswitch RBS | 28 | |
| BBa_K4605013 | Shine-Dalgarno sequence | 9 | |
| BBa_K4609058 | RBS | 23 | |
| BBa_K4611020 | RBS from bacteriophage T7 gene 10 | 23 | |
| BBa_K4613098 | RBS | 7 | |
| BBa_K4613099 | RBS(pQE80L) | 6 | |
| BBa_K4614109 | RBS for E.coli | 6 | |
| BBa_K4632013 | BBa_B0032X(medium) | 26 | |
| BBa_K4632021 | RBS32Z | 46 | |
| BBa_K4632023 | BBa_B0033 gain from literature | 24 | |
| BBa_K4633005 | RBS paraR - the RBS from the promoter for the araR gene from Bacillus subtilis 168 | 12 | |
| BBa_K4633007 | RBS paraE - the RBS from the promoter for the araE gene from Bacillus subtilis 168 | 14 | |
| BBa_K4634001 | RBS | 23 | |
| BBa_K4635009 | This is the RBS1 of Escherichia coli, which can bind to ribosomes and start the translation of downs | 6 | |
| BBa_K4635010 | This is the RBS2 of Escherichia coli, which can bind to ribosomes and start the translation of downs | 5 | |
| BBa_K4635011 | This is the RBS3 of Escherichia coli, which can bind to ribosomes and start the translation of downs | 5 | |
| BBa_K4635012 | This is the RBS4 of Escherichia coli, which can bind to ribosomes and start the translation of downs | 4 | |
| BBa_K4635013 | This is the RBS5 of Escherichia coli, which can bind to ribosomes and start the translation of downs | 4 | |
| BBa_K4643009 | Strong RBS | 12 | |
| BBa_K4645010 | a new ribosome-binding site | 34 | |
| BBa_K4645011 | a new ribosome-binding site 2 | 32 | |
| BBa_K4660004 | promotor-VbrR | 302 | |
| BBa_K4660005 | promotor-BlaI | 83 | |
| BBa_K4662000 | RBS initiation 20'000 | 28 | |
| BBa_K4662001 | RBS initiation 80'000 | 27 | |
| BBa_K4662002 | RBS initiation 140'000 | 27 | |
| BBa_K4662003 | RBS initiation 200'000 | 28 | |
| BBa_K4666023 | RBS-9K | 25 | |
| BBa_K4691001 | Modified RBS30 | 27 | |
| BBa_K4691002 | Modified RBS35 | 26 | |
| BBa_K4691003 | Modified RBS64 | 24 | |
| BBa_K4691004 | Modified RBS33 | 23 | |
| BBa_K4696005 | kink turns (k-turns) are RNA structural motifs that can be bound by a varieties of RNA-binding prote | 65 | |
| BBa_K4696008 | (IRES) Internal Ribosome Entry Site | 611 | |
| BBa_K4701230 | Synthetic RBS for tpaK | 25 | |
| BBa_K4701231 | Synthetic RBS for tphA2 | 25 | |
| BBa_K4701232 | Synthetic RBS for tphA3 | 25 | |
| BBa_K4701233 | Synthetic RBS for tphB | 25 | |
| BBa_K4701234 | Synthetic RBS for tphA1 | 25 | |
| BBa_K4705000 | This gene is designed in our system 1. With the function of N-acetyltransferase in lactobacillus pla | 149 | |
| BBa_K4706004 | 10bp Kozak | 6 | |
| BBa_K4711015 | alkB(RBS) | 25 | |
| BBa_K4711016 | alkG(RBS) | 28 | |
| BBa_K4711017 | alkT(RBS) | 31 | |
| BBa_K4711018 | GDH(RBS) | 6 | |
| BBa_K4719006 | RBS2 | 12 | |
| BBa_K4719007 | RBS3 | 12 | |
| BBa_K4719008 | RBS4 | 12 | |
| BBa_K4721010 | RBS to combine with PV10 promoter | 13 | |
| BBa_K4721013 | RBS to combine with Pbla-mut1T | 9 | |
| BBa_K4728015 | PhaJ (Hydratase) | ||
| BBa_K4729100 | RBS FL_A | 42 | |
| BBa_K4729101 | RBS T1_A | 32 | |
| BBa_K4729102 | RBS T3_A | 19 | |
| BBa_K4729103 | RBS FL_G | 42 | |
| BBa_K4729104 | RBS GGT | 15 | |
| BBa_K4729105 | RBS3 | 16 | |
| BBa_K4733001 | B. Subtilis RBS | 8 | |
| BBa_K4735001 | rbs sequence | 37 | |
| BBa_K4735009 | rbs for sfgfp | 52 | |
| BBa_K4755002 | pET22B(+) RBS | 28 | |
| BBa_K4757002 | synthetic RBS to maximize translation of Gel.Exp.-LCC | 27 | |
| BBa_K4757003 | synthetic RBS to maximize translation of mKate2 | 43 | |
| BBa_K4757004 | synthetic RBS to maximize translation of AlkB | 49 | |
| BBa_K4757005 | synthetic RBS to maximize translation of SPpstu-FAST-PETase | 42 | |
| BBa_K4769606 | T7 g10-L RBS | ||
| BBa_K4769607 | T7 g10-L RBS | 21 | |
| BBa_K4790079 | RBS | 23 | |
| BBa_K4796003 | Strong 5'UTR for Vibrio natriegens (BBa_K3187014 derivative) | ||
| BBa_K4796004 | Synthetic strong 5'UTR for Vibrio natriegens | ||
| BBa_K4796005 | Strong 5'UTR for Vibrio natriegens (BBa_K3187014 derivative) | 49 | |
| BBa_K4807029 | RBS from pETDuet-1 | 43 | |
| BBa_K4811000 | repressible RBS (BRA) | 24 | |
| BBa_K4813019 | Modified RBS for T7 promoter | 41 | |
| BBa_K4813021 | RBS for wild-type hxlAB operon transcription | 8 | |
| BBa_K4814012 | Strong RBS | 20 | |
| BBa_K4818106 | RBS | 5 | |
| BBa_K4822995 | sae_RBS for BBa_K4822005 | 19 | |
| BBa_K4831004 | RBS_S3* (Thiel et al. 2018) | 54 | |
| BBa_K4831005 | RBS_S5* (Thiel et al. 2018) | 54 | |
| BBa_K4831008 | RBS_S4* (Thiel et al.) | 54 | |
| BBa_K4833020 | This gene transcribes an mRNA sequence which is a ribosome binding site. | 14 | |
| BBa_K4841014 | PesaS RBS | 27 | |
| BBa_K4877018 | minimal 6bp Kozak sequence | 6 | |
| BBa_K4891022 | RBS | 5 | |
| BBa_K4895998 | 5' UTR from genomic E.coli | 11 | |
| BBa_K4895999 | 5 | 213 | |
| BBa_K4902007 | Modifed Shine-Dalgrano RBS | 16 | |
| BBa_K4907041 | T7 RBS | 29 | |
| BBa_K4911000 | Modified sequence of a part BBa_J433006 with modified BsaI 3 | 86 | |
| BBa_K4911001 | Modified sequence of a part BBa_J433003 with modified BsaI 3' fusion site AATG | 50 | |
| BBa_K4916002 | trpL Shine-Dalgarno (RBS) sequence | 13 | |
| BBa_K4927048 | RBS | 23 | |
| BBa_K4927056 | RBS | 6 | |
| BBa_K4930009 | Shine-Dalgarno-Sequence | 7 | |
| BBa_K4931005 | RBS5 | 37 | |
| BBa_K4931006 | RBS6 | 37 | |
| BBa_K4931007 | RBS7 | 37 | |
| BBa_K4931008 | RBS8 | 37 | |
| BBa_K4931009 | RBS9 | 37 | |
| BBa_K4931010 | RBS10 | 37 | |
| BBa_K4931011 | RBS11 | 44 | |
| BBa_K4931012 | RBS12 | 44 | |
| BBa_K4931013 | RBS13 | 44 | |
| BBa_K4931015 | RBS15 | 44 | |
| BBa_K4931016 | RBS16 | 44 | |
| BBa_K4934005 | RBS | 18 | |
| BBa_K4934006 | RBS | 18 | |
| BBa_K4934007 | RBS | 18 | |
| BBa_K4934008 | RBS | 18 | |
| BBa_K4934009 | RBS | 18 | |
| BBa_K4934010 | RBS | 18 | |
| BBa_K4934011 | RBS | 18 | |
| BBa_K4937012 | Kozak sequence | 6 | |
| BBa_K4940004 | RBS in pCDFDuet-1 | 6 | |
| BBa_K4940006 | RBS in pQE-80L-Kan | 12 | |
| BBa_K4940007 | RBS in pET-28a | 7 | |
| BBa_K4942003 | PluxCDABEG | 377 | |
| BBa_K4943058 | Riboswitch with Cas9 nuclease (codon optimized) | 1039 | |
| BBa_K4947007 | RBS for At4CL1, expressed in E. coli | 35 | |
| BBa_K4947008 | RBS for Gu4CL1, expressed in E. coli | 35 | |
| BBa_K4947009 | RBS for GmCHS8, expressed in E. coli | 35 | |
| BBa_K4947010 | RBS for GmCHR5, expressed in E. coli | 35 | |
| BBa_K4947011 | RBS for GuCHR1, expressed in E. coli | 35 | |
| BBa_K4947012 | RBS for GmCHIB2, expressed in E. coli | 35 | |
| BBa_K4947013 | RBS for GuCHI1, expressed in E. coli | 35 | |
| BBa_K4947014 | RBS for MsCHI1, expressed in E. coli | 35 | |
| BBa_K4947015 | RBS for CrCPR, expressed in E. coli | 34 | |
| BBa_K4947016 | RBS for GmCPR, expressed in E. coli | 35 | |
| BBa_K4947017 | RBS for Ge2-HIS, expressed in E. coli | 35 | |
| BBa_K4947018 | RBS for TpIFS, expressed in E. coli | 35 | |
| BBa_K4947019 | RBS for GmHID, expressed in E. coli | 35 | |
| BBa_K4948004 | creRBS | 19 | |
| BBa_K4948016 | Strong ribosome binding site from pET vector | 23 | |
| BBa_K4951000 | KanR Toehold 1 | 167 | |
| BBa_K4951001 | KanR Toehold 2 | 167 | |
| BBa_K4951002 | KanR Toehold 3 | 167 | |
| BBa_K4951003 | KanR Toehold 4 | 167 | |
| BBa_K4951004 | KanR Toehold 5 | 167 | |
| BBa_K4951005 | KanR Toehold 6 | 167 | |
| BBa_K4951006 | KanR Toehold 7 | 167 | |
| BBa_K4951007 | KanR Toehold 8 | 167 | |
| BBa_K4951008 | KanR Toehold 9 | 167 | |
| BBa_K4951009 | KanR Toehold 10 | 167 | |
| BBa_K4951010 | AmpR Toehold 1 | 158 | |
| BBa_K4951011 | AmpR Toehold 2 | 158 | |
| BBa_K4951012 | AmpR Toehold 3 | 158 | |
| BBa_K4951013 | AmpR Toehold 4 | 158 | |
| BBa_K4951014 | KanR Riboswitch 1 | 176 | |
| BBa_K4951015 | KanR Riboswitch 2 | 176 | |
| BBa_K4951016 | KanR Riboswitch 3 | 176 | |
| BBa_K4951017 | KanR Riboswitch 4 | 176 | |
| BBa_K4951018 | KanR Riboswitch 5 | 176 | |
| BBa_K4951019 | KanR Riboswitch 6 | 176 | |
| BBa_K4951020 | KanR Riboswitch 7 | 176 | |
| BBa_K4951021 | KanR Riboswitch 8 | 176 | |
| BBa_K4951022 | KanR Riboswitch 9 | 176 | |
| BBa_K4951023 | KanR Riboswitch 10 | 176 | |
| BBa_K4951024 | KanR Riboswitch 11 | 176 | |
| BBa_K4951025 | KanR Riboswitch 12 | 176 | |
| BBa_K4951026 | KanR Riboswitch 13 | 159 | |
| BBa_K4986001 | Suicide system: temperature-controlled promoter +SRRZ cutting gene | 3021 | |
| BBa_K4998024 | RBS for VapC15 gene | 12 | |
| BBa_K5038000 | RBS | 8 | |
| BBa_K5044012 | RBS | 23 | |
| BBa_K5044037 | pQQC7_RBS | 12 | |
| BBa_K5044101 | taccD 5 | 180 | |
| BBa_K5044108 | CrTrbcl | 450 | |
| BBa_K5044304 | CrTrbcl | 450 | |
| BBa_K5048063 | RBS(On pET-28a backbone) | 23 | |
| BBa_K5048064 | RBS(On pBAD backbone) | 6 | |
| BBa_K5061021 | Synthetic RBS designed for M13 gI | 30 | |
| BBa_K5061022 | Synthetic RBS designed for M13 gIII | 22 | |
| BBa_K5061023 | Synthetic RBS designed for M13 gVI | 22 | |
| BBa_K5061024 | Synthetic RBS designed for XylS | 26 | |
| BBa_K5062038 | Internal Ribosome Entry Site (IRES) | 568 | |
| BBa_K5076113 | vioA RBS | 6 | |
| BBa_K5076203 | viaA RBS | 6 | |
| BBa_K5084006 | Thiosulfate biosensor-A-RBS of thsS | 34 | |
| BBa_K5084007 | Thiosulfate biosensor-A-RBS of thsR | 33 | |
| BBa_K5084008 | Thiosulfate biosensor-B-RBS of thsS | 28 | |
| BBa_K5084009 | Thiosulfate biosensor-B-RBS of thsS | 33 | |
| BBa_K5097012 | RBS BBa_B0034 with spacer nucleotides | 26 | |
| BBa_K5108006 | aprA RBS from Pseudomonas protegens (CHA0) | 24 | |
| BBa_K5108007 | hcnA RBS from Pseudomonas protegens (CHA0) | 25 | |
| BBa_K5108008 | hcnA RBS from Pseudomonas aeruginosa (PAO1) | 25 | |
| BBa_K5117000 | BsRBS | 15 | |
| BBa_K5133001 | Ribosome binding site (RBS, from plasmid pJL1) | 17 | |
| BBa_K5136045 | SD7 | 7 | |
| BBa_K5136049 | SD17 | 7 | |
| BBa_K5160001 | Short Description | ||
| BBa_K5166022 | UreA RBS1 | 25 | |
| BBa_K5166023 | UreB RBS2 | 25 | |
| BBa_K5166024 | UreC RBS3 | 25 | |
| BBa_K5166025 | UreE RBS4 | 25 | |
| BBa_K5166026 | UreF RBS5 | 25 | |
| BBa_K5166027 | UreG RBS6 | 25 | |
| BBa_K5166028 | UreD RBS7 | 25 | |
| BBa_K5175014 | RBS for T7 promoter | 12 | |
| BBa_K5211004 | The ribosome-binding site is a sequence on the mRNA that is located upstream of the start codon (usu | 24 | |
| BBa_K5211005 | Optimized ribosome-binding site sequences. | 26 | |
| BBa_K5226019 | RBS for TD-gcvT | 35 | |
| BBa_K5226020 | RBS for TD-gcvH | 35 | |
| BBa_K5226021 | RBS for TD-gcvP | 35 | |
| BBa_K5226022 | RBS for TD-sdaA | 35 | |
| BBa_K5226023 | RBS for Vib-folD | 35 | |
| BBa_K5226024 | RBS for Vib-gcvT | 35 | |
| BBa_K5226025 | RBS for Vib-gcvH | 35 | |
| BBa_K5226026 | RBS for Vib-gcvP | 35 | |
| BBa_K5226027 | RBS for Vib-sdaA | 35 | |
| BBa_K5226035 | RBS for 4hbD | 35 | |
| BBa_K5226036 | RBS for sucD | 35 | |
| BBa_K5226040 | RBS for cysNC | 35 | |
| BBa_K5246000 | RBS_1MCS_HB | 6 | |
| BBa_K5246053 | RBS | 6 | |
| BBa_K5248013 | RBS | 23 | |
| BBa_K5254009 | RBS Derived using Geneious Prime | 11 | |
| BBa_K5269034 | SD-sequence | 16 | |
| BBa_K5270006 | RBS in L.reuteri | 6 | |
| BBa_K5280410 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280411 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280412 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280413 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280414 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280415 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280416 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280417 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280418 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280419 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280420 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280421 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280422 | A RNA thermosensor that turns on protein translation at high temperatures. | 71 | |
| BBa_K5280423 | A RNA thermosensor that turns on protein translation at high temperatures. | 73 | |
| BBa_K5280424 | A RNA thermosensor that turns on protein translation at high temperatures. | 73 | |
| BBa_K5280425 | A RNA thermosensor that turns on protein translation at high temperatures. | 73 | |
| BBa_K5280426 | A RNA thermosensor that turns on protein translation at high temperatures. | 73 | |
| BBa_K5280427 | A RNA thermosensor that turns on protein translation at high temperatures. | 100 | |
| BBa_K5280428 | A RNA thermosensor that turns on protein translation at high temperatures. | 103 | |
| BBa_K5280429 | A RNA thermosensor that turns on protein translation at high temperatures. | 103 | |
| BBa_K5280430 | A RNA thermosensor that turns on protein translation at high temperatures. | 103 | |
| BBa_K5280431 | A RNA thermosensor that turns on protein translation at high temperatures. | 107 | |
| BBa_K5282006 | RBS | 13 | |
| BBa_K5282009 | RBS | 13 | |
| BBa_K5282010 | TfCa CDS | 1530 | |
| BBa_K5291012 | BCD1 bicistronic design | 85 | |
| BBa_K5291013 | BCD2 bicistronic design | 83 | |
| BBa_K5291014 | BCD5 bicistronic design | 88 | |
| BBa_K5293010 | RBS T7 | 30 | |
| BBa_K5293021 | pHREAC RBS | 33 | |
| BBa_K5300003 | RBS1 | 40 | |
| BBa_K5300006 | RBS2 | 40 | |
| BBa_K5300008 | RBS3 | 40 | |
| BBa_K5300010 | RBS4 | 40 | |
| BBa_K5300012 | Ribosome binding site of VapC | 22 | |
| BBa_K5303057 | RBS | 6 | |
| BBa_K5303101 | SRBS | 21 | |
| BBa_K5311004 | Strong synthetic RBS | 29 | |
| BBa_K5327000 | AVBBAA | 1065 | |
| BBa_K5348001 | B0034-mutant 1 | 15 | |
| BBa_K5348002 | B0034-mutant 2 | 12 | |
| BBa_K5348003 | B0034-mutant 3 | 12 | |
| BBa_K5357000 | E. coli Ribosome Binding Site (RBS) | 23 | |
| BBa_K5359000 | SYFP2 RBS | 39 | |
| BBa_K5366070 | RBS | 6 | |
| BBa_K5366072 | RBS (J23119 promoter) | 23 | |
| BBa_K5369105 | Ribosome binding site | 6 | |
| BBa_K5377001 | ribA RBS | 27 | |
| BBa_K5377003 | ribB RBS | 28 | |
| BBa_K5377005 | ribD RBS | 26 | |
| BBa_K5377007 | ribE RBS | 33 | |
| BBa_K5377009 | ribC RBS | 35 | |
| BBa_K5377504 | efficient RBS | 23 | |
| BBa_K5378016 | RBS | 26 | |
| BBa_K5382102 | RBS | 23 | |
| BBa_K5398004 | RBS (before 5' intron) | 15 | |
| BBa_K5422002 | RBS with spacers | 30 | |
| BBa_K5427000 | RBS 1_12000 | 14 | |
| BBa_K5427005 | RBS 2_220 | 14 | |
| BBa_K5427006 | RBS 3_126 | 32 | |
| BBa_K5427007 | RBS 4_59 | 33 | |
| BBa_K5431009 | RBS (MP6) | 28 | |
| BBa_K5431013 | RBS (from T7) | 69 | |
| BBa_K5432012 | RBS(Ribosome binding site) | 43 | |
| BBa_K5436002 | Optimized RBS for BIND-System | 24 | |
| BBa_K5436005 | Optimized RBS for BIND-System | 24 | |
| BBa_K5441008 | RBS | 12 | |
| BBa_K545005 | fha | 0.1 | 137 |
| BBa_K545006 | mag | 0.04 | 32 |
| BBa_K545666 | rpos leader sequence | 223 | |
| BBa_K5487103 | RBS | 23 | |
| BBa_K5487117 | RBS | 23 | |
| BBa_K5490016 | RBS | 30 | |
| BBa_K5490030 | IRES2 | 587 | |
| BBa_K5490031 | kozak for casrx | 24 | |
| BBa_K5498012 | RBS | 67 | |
| BBa_K566037 | RBS.3 (Inverted) | 13 | |
| BBa_K606061 | SpoVG Ribosome Binding Site (RBS) for B. subtilis | 12 | |
| BBa_K619892 | mutated prfA thermosensor from listeria | 140 | |
| BBa_K624012 | RBS from Pmsp3 | 31 | |
| BBa_K624013 | RBS (6 bps truncated) from Pmsp3 | 22 | |
| BBa_K641001 | rbs for ffGFP | 30 | |
| BBa_K641004 | rbs for ToxR | 25 | |
| BBa_K737020 | Cyanobacteria gene ribosome binding site | 5 | |
| BBa_K737033 | galK is the leader sequence of galactokinase (GalK) | 127 | |
| BBa_K780001 | Strong RBS for Bacillus Subtilis | 16 | |
| BBa_K780002 | Strong RBS for Bacillus Subtilis | 16 | |
| BBa_K783047 | This is a MoClo converted version of BBa_B0030 | 15 | |
| BBa_K783048 | This is a MoClo converted version of BBa_B0031 | 14 | |
| BBa_K783049 | This is a MoClo converted version of BBa_B0032 | 13 | |
| BBa_K783050 | This is a MoClo converted version of BBa_B0033 | 11 | |
| BBa_K783051 | This is a MoClo converted version of BBa_B0034 | 12 | |
| BBa_K784007 | Spacer + RBS from theophylline riboswitch | 46 | |
| BBa_K784012 | Spacer2 + RBS from theophylline riboswitch | 51 | |
| BBa_K792001 | Kozak sequence from yeast α-factor mating pheromone (MFα1) | 21 | |
| BBa_K809008 | RBS sequence of yeast mitochondrial gene AI1 | 54 | |
| BBa_K809009 | RBS sequence of yeast mitochondrial gene OLI1 | 114 | |
| BBa_K812053 | RBS B0034 with GoldenBrick sequence | 47 | |
| BBa_K812058 | A GoldenBricked-medium strenght RBS J61107 | 48 | |
| BBa_K812059 | A GoldenBricked version of week RBS J61117 | 48 | |
| BBa_K821009 | Orthogonal RBS 1 | 6 | |
| BBa_K879021 | rbs based on B0034 | 26 | |
| BBa_K879022 | Rbs 2 | 29 | |
| BBa_K879023 | Rbs 3 | 29 | |
| BBa_K879024 | rbs 4 | 29 | |
| BBa_K879025 | Rbs 5 | 29 | |
| BBa_K879026 | Rbs 6 | 29 | |
| BBa_K892005 | Strong RBS | 16 | |
| BBa_K896599 | Strophurus rankini voucher AMS R140490 NADH dehydrogenase subunit 4 (ND4) gene, partial cds; tRNA-Hi | 435 | |
| BBa_K899009 | Doubble ribosomal binding site (FFT) (shine-dalgarno and kozak sequence) | 13 | |
| BBa_K899010 | Doubble ribosomal binding site (SST) (shine-dalgarno and kozak sequence) | 13 | |
| BBa_K923005 | Ribosome Binding Site in pET14b plasmid | 5 | |
| BBa_M1105 | Pyrene degradation enzyme RBS | 11 | |
| BBa_M11402 | 5' UTR and RBS of psbA2 gene in Synechocystis sp. PCC 6803 | 49 | |
| BBa_M13501 | M13K07 gene I RBS | 16 | |
| BBa_M13502 | M13KO7 gene II RBS | 16 | |
| BBa_M13503 | M13K07 gene III RBS | 16 | |
| BBa_M13504 | M13K07 gene IV RBS | 16 | |
| BBa_M13505 | M13KO7 gene V RBS | 16 | |
| BBa_M13506 | M13K07 gene VI RBS | 16 | |
| BBa_M13507 | M13KO7 gene VII RBS | 16 | |
| BBa_M13508 | M13K07 gene VIII RBS | 16 | |
| BBa_M13509 | M13Ko7 gene IX RBS | 16 | |
| BBa_M13510 | M13KO7 gene X RBS | 16 | |
| BBa_M13511 | M13K07 gene XI RBS | 15 | |
| BBa_M1694 | Ribosome binding site | 14 | |
| BBa_M1782 | Synechocystis RBS Site | 10 | |
| BBa_M36000 | 5' Bicistronic UTR (weak), does not include ATG start | 47 | |
| BBa_M36001 | 5' Bicistronic UTR (medium), does not include ATG start | 47 | |
| BBa_M36009 | 5' Bicistronic UTR (strong), does not include ATG start | 47 | |
| BBa_M36034 | BCD02 - Bicistronic Design 02 | 85 | |
| BBa_M36068 | Adapted fnr binding site | 130 | |
| BBa_M36070 | Bicistronic RBS - BIOFAB ID: apFAB681 (BCD1) | 88 | |
| BBa_M36120 | 5' Bicistronic UTR (strong) contains ATG start Codon. | 88 | |
| BBa_M36204 | Bicistronic RBS (BD14) | 85 | |
| BBa_M36236 | 5' Bicistronic UTR (no ATG) | 85 | |
| BBa_M36237 | 5' Bicistronic UTR (no ATG) | 85 | |
| BBa_M36269 | RBS: modified BBa_B0034 | 15 | |
| BBa_M36282 | 5' Bicistronic UTR, includes ATG start codon | 88 | |
| BBa_M36288 | medium 5' Bicistronic UTR (BD21) | 88 | |
| BBa_M36289 | medium 5' Bicistronic UTR (BD8) | 88 | |
| BBa_M36293 | 5' Bicistronic UTR, includes ATG start | 88 | |
| BBa_M36361 | 5' Bicistronic UTR (apFAB866, GENE10_BCD2), No ATG Start | 83 | |
| BBa_M36405 | Bicistronic RBS (medium) pFAB1646 | 88 | |
| BBa_M36450 | Bicistronic RBS - pFAB1692 | 85 | |
| BBa_M36452 | RBS - MCD 6 | 29 | |
| BBa_M36454 | MCD 20 | 30 | |
| BBa_M36516 | Bicistronic RBS #5 | 88 | |
| BBa_M36521 | strong 5' bicistronic UTR, with ATG at end | 86 | |
| BBa_M36542 | BCD22 - RBS | 88 | |
| BBa_M36556 | 5' Bicistronic UTR (medium), does not include ATG start | 85 | |
| BBa_M36559 | RBS and Spacer | 19 | |
| BBa_M36583 | Strong RBS - BBa_J61100 | 28 | |
| BBa_M36595 | Bicistronic design RBS | 87 | |
| BBa_M36724 | 5' Bicistronic UTR (strong), does not include ATG start | 85 | |
| BBa_M36771 | Strong RBS from DNA 2.0 Gene Designer | 13 | |
| BBa_M36781 | BCD13 | 92 | |
| BBa_M36804 | pRBS-SD1+6A | 13 | |
| BBa_M36913 | takes PoPs input | 88 | |
| BBa_M36917 | Zinc Promoter (ZntR regulated) with own RBS | 91 | |
| BBa_M36925 | Bicistronic RBS (medium) apFAB682 | 88 | |
| BBa_M45193 | E. coli RBS | 14 | |
| BBa_M45214 | Physcomitrella patens ribosome binding site | 207 | |
| BBa_M45406 | minimal TATA Box | 64 | |
| BBa_M50007 | U1 Thermosensitive 5'-UTR (includes RBS) | 38 | |
| BBa_M50008 | U10 Thermosensitive 5'-UTR (includes RBS) | 39 | |
| BBa_M50019 | Strong ribosome binding site | 15 | |
| BBa_M50024 | RBS for Gold detection plasmid | 10 | |
| BBa_M50062 | Strong RBS | 13 | |
| BBa_M50080 | Strong RBS | 13 | |
| BBa_M50422 | Strong RBS | 13 | |
| BBa_Z0261 | Strong T7.2 RBS | 20 | |
| BBa_Z0262 | Medium strength T7.2 RBS | 20 | |