Part:BBa_K3585000
the RBS in pMTL83151
A Ribosome Binding Site.
Characterization by mathematical modeling by AFCM-Egypt 2022
We are using mathematical modeling to detect the increased level of phenylalanine (phe) in phenylketonuria patients in our diagnostic platform. It depends on a whole cell-based biosensor through a cascade of reactions to finally end by formation of β-galactosidase that turns the color into blue once bound to its substrate (X-gal) as mentioned in figure (2) and graph (1).
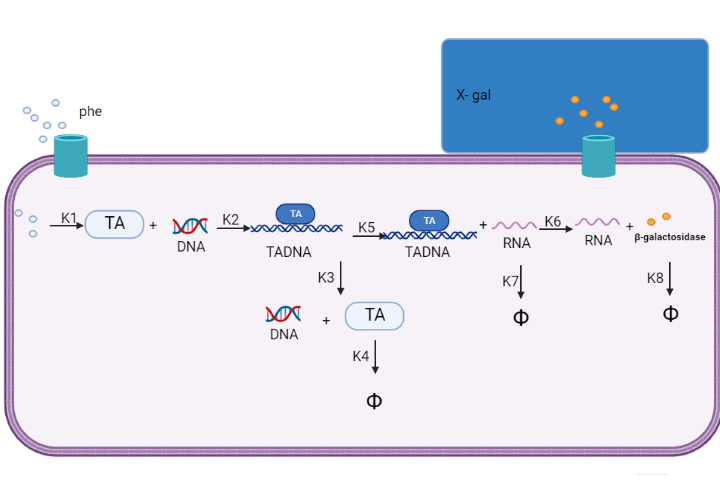
Figure (2) represents the cascade of reactions in whole cell-based biosensor model.
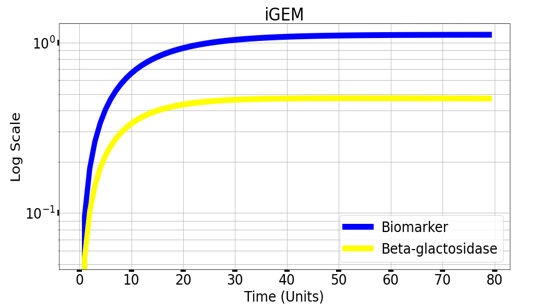
Graph(1) illustrates a direct relation between biomarker and beta-galactosidase ,so as the biomarker increases, the released amount of beta-galactosidase increases till it reaches constant value after about 30 time units. Therefore, the maximum amount of the biomarker releases the maximum amount of beta-galactosidase.
Experimental Characterization by AFCM-Egypt 2022 team
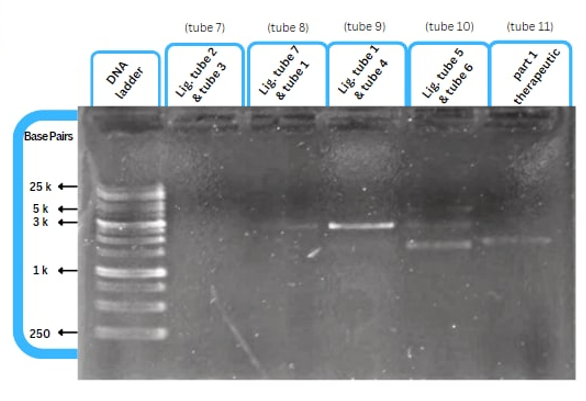
This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane 6 (the last one). The running part (ordered from IDT) included T7P - TyrR RBS - TyrR - TyrPromoter.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |