Part:BBa_K4294005
RBS Ath5
A synthetic ribosome binding site for prokaryotic protein expression, built with the guidance of the RBS calculator and characterized by our team.
Measurment
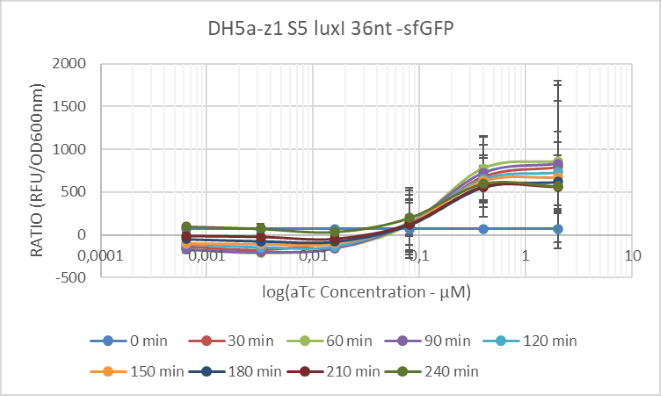
We characterized this RBS in the genetic context of luxI. We built a fusion of the first 36 nucleotides of the LuxI synthase with sfGFP to characterize the relative strength of our deployed RBS in the context of the LuxI coding sequence. To quantify this output we measured the fluorescence using microplate reader FlexStation3 (Molecular Devices) with excitation wavelength 485nm and emission wavelength 510nm. We conducted measurements in different time points after the induction with aTc, using different concentrations of the inducer.
'Figure 1 : Characterization of Synthetic 5 RBS in the genetic context of luxI'
'Figure 2 : Comparison of all RBS characterized by our team'
References
[1] Reis, A. C., & Salis, H. M. (2020). An Automated Model Test System for Systematic Development and Improvement of Gene Expression Models. ACS synthetic biology, 9(11), 3145–3156. https://doi.org/10.1021/acssynbio.0c00394
[2] Salis, H. M., Mirsky, E. A., & Voigt, C. A. (2009). Automated design of synthetic ribosome binding sites to control protein expression. Nature biotechnology, 27(10), 946–950. https://doi.org/10.1038/nbt.1568
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |