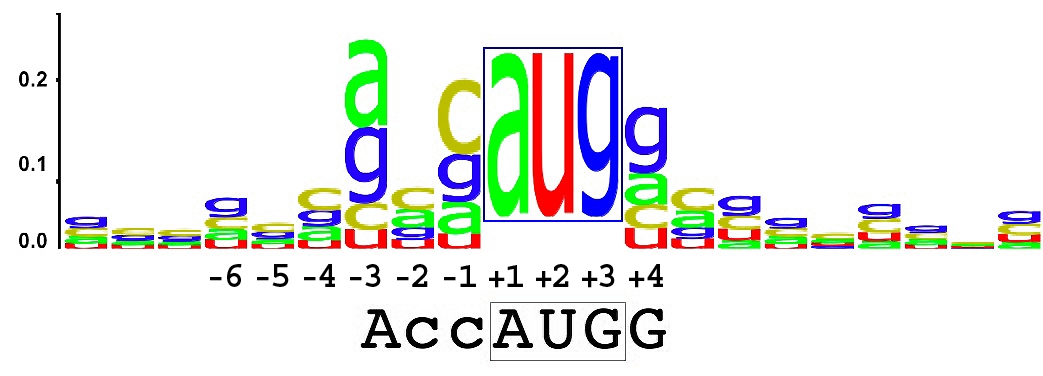Yeast
S. cerevisiae, a species of budding yeast, is a convenient chassis for engineered biological systems for several reasons.
- It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology.
- As a single celled organism S. cerevisiae is small with a short generation time (doubling time 1.5–2 hours at 30°C) and can be easily cultured.
- S. cerevisiae can be transformed allowing for either the addition of new genes or deletion through homologous recombination. Furthermore, The ability to grow S. cerevisiae as a haploid simplifies the creation of gene knockouts strains.
- As a eukaryote, S. cerevisiae shares the complex internal cell structure of plants and animals without the high percentage of non-coding DNA that can confound research in higher eukaryotes.
As an aside, it is perhaps the most useful yeast owing to its use since ancient times in baking and brewing.
| Plasmid backbones (?) | Promoters (?) | Kozak sequences (?) | Protein domains (?) | Protein coding sequences (?) | Translational units (?) | Terminators (?) |
| Help: Want to know more about Yeast? See the help pages for more information. |
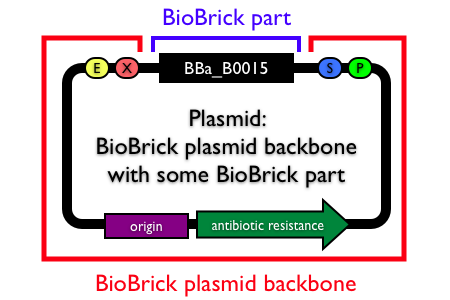
Plasmid backbones
Plasmids are circular, double-stranded DNA molecules typically containing a few thousand base pairs that replicate within the cell independently of the chromosomal DNA. Plasmid DNA is easily purified from cells, manipulated using common lab techniques and incorporated into cells. Most BioBrick parts in the Registry are maintained and propagated on plasmids. Thus, construction of BioBrick parts, devices and systems usually requires working with plasmids.
Note: In the Registry, plasmids are made up of two distinct components:
- the BioBrick part, device or system that is located in the BioBrick cloning site, between (and excluding) the BioBrick prefix and suffix.
- the plasmid backbone which propagates the BioBrick part. The plasmid backbone is defined as the sequence beginning with the BioBrick suffix, including the replication origin and antibiotic resistance marker, and ending with the BioBrick prefix. [Note that the plasmid backbone itself can be composed of BioBrick parts.]
Many BioBrick parts in the Registry are maintained on more than one plasmid backbone!
Note that most of these plasmid backbones comply with the Lim lab AarI part assembly standard. See [http://2008.igem.org/Everything_you_ever_wanted_to_know_about_AarI the 2008 UCSF iGEM team wiki] for more details.
| Name | Description | Resistance | Replicon | Copy number | Chassis | Length |
|---|---|---|---|---|---|---|
| BBa_J63010 | Protein fusion vector (Silver lab standard) | A | 3266 | |||
| BBa_K106006 | AarI AD acceptor vector (pRS315, Adh1P, Adh1t) | 7650 | ||||
| BBa_K106014 | AarI AD acceptor vector (pRS315, Cyc1P, Adh1t) | 6397 | ||||
| BBa_K106670 | AarI AD acceptor, pRS315, 8X LexAOPs Cyc1P, Adht1 | 6559 | ||||
| BBa_K106672 | AarI AD acceptor vector (pRS305, Gal1P, Adh1t) | 6638 | ||||
| BBa_K106693 | AarI A!D acceptor vector (pRS315, Cyc1P, Adh1t) | 7650 | ||||
| BBa_K106697 | AarI AD acceptor vector (pRS315, Cyc1P, Adh1t-8XLexA Ops) | 6559 | ||||
| BBa_K106698 | AarI AD acceptor vector (pRS315, 8X LexAOps Fig1P, Adh1t) | 7364 | ||||
| BBa_K1680014 | pRS313 yeast shuttle vector with Biobrick MCS | 4928 | ||||
| BBa_K1680015 | pRS315 yeast shuttle vector with Biobrick MCS | 5979 | ||||
| BBa_K1680016 | pRS316 yeast shuttle vector with Biobrick MCS | 4848 | ||||
| BBa_K319043 | ADE4 targeting vector | 2508 | ||||
| BBa_K394001 | Plasmid for Chromosomal Integration in Yeast at His3 | 3457 | ||||
| BBa_K394002 | Plasmid for Chromosomal Integration in Yeast at Ura3 | 3458 | ||||
| BBa_K4188001 | pSB3CY_aeBlue shuttle vector backbone for Saccharomyces cerevisiae | 6171 | ||||
| BBa_K4706000 | pSB1C30YIpHR-HO Chromosomal integration at HO loci | chloramphenicol | High copy for E. coli; Chromosomal integrated in S. cerevisae | E. coli for cloning; S. cerevisiae for Homologous integration | 3070 | |
| BBa_K555009 | pYES2 - galactose-inducible expression vector for yeast | 5856 | ||||
| BBa_K801000 | pTUM100 yeast shuttle vector based on pYES2 | 4923 | ||||
| BBa_K801001 | pTUM101 yeast shuttle vector with pTEF1 promoter | 5315 | ||||
| BBa_K801002 | pTUM102 yeast shuttle vector with pTEF2 promoter | 5514 | ||||
| BBa_K801003 | pTUM103 yeast shuttle vector with pADH1 promoter | 5585 | ||||
| BBa_K801004 | pTUM104 yeast shuttle vector with GAL1 promoter | 5837 |

|

|
Sergio Peisajovich and Andrew Horowitz, from Wendell Lim's lab, developed several of the yeast plasmid backbones as an instructor of the 2008 UCSF iGEM team. |
Promoters
A promoter is a DNA sequence that can recruit transcriptional machinery and lead to transcription of the downstream DNA sequence. The specific sequence of the promoter determines the strength of the promoter (a strong promoter leads to a high rate of transcription initiation).
In addition to sequences that "promote" transcription, a promoter may include additional sequences known as operators that control the strength of the promoter. For example, a promoter may include a binding site for a protein that attracts or obstructs the RNAP binding to the promoter. The presence or absence of the protein will affect the strength of the promoter. Such a promoter is known as a regulated promoter.
Constitutive
All the promoters on this page are yeast promoters that are constitutive meaning that their activity is dependent on the availability of RNA polymerase holoenzyme, but is not affected by any transcriptional regulators. If you find any promoters on this page that you know to be regulated by a particular transcription factor, please let us know, or re-categorize the part yourself!
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_I766555 | pCyc (Medium) Promoter | . . . acaaacacaaatacacacactaaattaata | 244 | 3344 | Not in stock | ||
| BBa_I766556 | pAdh (Strong) Promoter | . . . ccaagcatacaatcaactatctcatataca | 1501 | 3301 | Not in stock | ||
| BBa_I766557 | pSte5 (Weak) Promoter | . . . gatacaggatacagcggaaacaacttttaa | 601 | 3594 | Not in stock | ||
| BBa_J63005 | yeast ADH1 promoter | . . . tttcaagctataccaagcatacaatcaact | 1445 | 7638 | It's complicated | ||
| BBa_K105027 | cyc100 minimal promoter | . . . cctttgcagcataaattactatacttctat | 103 | 1876 | It's complicated | ||
| BBa_K105028 | cyc70 minimal promoter | . . . cctttgcagcataaattactatacttctat | 103 | 1833 | It's complicated | ||
| BBa_K105029 | cyc43 minimal promoter | . . . cctttgcagcataaattactatacttctat | 103 | 1832 | It's complicated | ||
| BBa_K105030 | cyc28 minimal promoter | . . . cctttgcagcataaattactatacttctat | 103 | 1832 | It's complicated | ||
| BBa_K105031 | cyc16 minimal promoter | . . . cctttgcagcataaattactatacttctat | 103 | 1832 | It's complicated | ||
| BBa_K122000 | pPGK1 | . . . ttatctactttttacaacaaatataaaaca | 1497 | 6078 | It's complicated | ||
| BBa_K124000 | pCYC Yeast Promoter | . . . acaaacacaaatacacacactaaattaata | 288 | 2076 | Not in stock | ||
| BBa_K124002 | Yeast GPD (TDH3) Promoter | . . . gtttcgaataaacacacataaacaaacaaa | 681 | 4340 | Not in stock | ||
| BBa_K319005 | yeast mid-length ADH1 promoter | . . . ccaagcatacaatcaactatctcatataca | 720 | 3236 | It's complicated | ||
| BBa_M31201 | Yeast CLB1 promoter region, G2/M cell cycle specific | . . . accatcaaaggaagctttaatcttctcata | 500 | 1858 | Not in stock |
Positively regulated
All the promoters on this page are yeast promoters that are positively regulated meaning that increased levels of at least one transcription factor (other than the sigma factor) will increase the activity of these promoters.
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_K392001 | yeast ENO2 promoter | . . . aagcaactaatactataacatacaataata | 598 | 2244 | It's complicated | ||
| BBa_K106699 | Gal1 Promoter | . . . aaagtaagaatttttgaaaattcaatataa | 686 | 1216 | Not in stock | ||
| BBa_K110015 | A-Cell Promoter MFA1 (RtL) | . . . cttcatatataaaccgccagaaatgaatta | 436 | 1659 | Not in stock | ||
| BBa_K110014 | A-Cell Promoter MFA2 (backwards) | . . . atcttcatacaacaataactaccaacctta | 550 | 1304 | Not in stock | ||
| BBa_K110004 | Alpha-Cell Promoter Ste3 | . . . gggagccagaacgcttctggtggtgtaaat | 501 | 1372 | Not in stock | ||
| BBa_J24813 | URA3 Promoter from S. cerevisiae | . . . gcacagacttagattggtatatatacgcat | 137 | 1431 | Not in stock | ||
| BBa_K4706014 | Yeast Gal1 Kozak partly | . . . gtcaaggaggaaactagacccgccgccacc | 543 | ||||
| BBa_K4365000 | FIG1 inducible promoter | . . . aaacaaccaaccaaaaaaaaaaaaaaaaaa | 1000 | ||||
| BBa_K2601000 | Promoter-Tet07(Saccharomyces cerevisiae) | . . . cacacactaaattacccggatcaattcggg | 724 | 1597 | It's complicated | ||
| BBa_K2294007 | Synthtic minimal Galactose induced promoter + Kozak sequence | . . . atcgaaaaaagtctccatggtggcggcggg | 160 | 6028 | It's complicated | ||
| BBa_K753000 | Yeast FIG1 promoter | . . . aaacaaacaaacaaaaaaaaaaaaaaaaaa | 450 | 2409 | It's complicated | ||
| BBa_K586000 | Cup-1 Heavy metal sensor | . . . accaagaagtcatgctgctctgggaaatga | 186 | 1888 | It's complicated | ||
| BBa_J63006 | yeast GAL1 promoter | . . . gaggaaactagacccgccgccaccatggag | 549 | 11798 | In stock | ||
| BBa_K284003 | Partial DLD Promoter from Kluyveromyces lactis | . . . aagtgcaagaaagaccagaaacgcaactca | 463 | 1735 | It's complicated | ||
| BBa_K284002 | JEN1 Promoter from Kluyveromyces lactis | . . . gagtaaccaaaaccaaaacagatttcaacc | 998 | 1654 | It's complicated | ||
| BBa_K165041 | Zif268-HIV binding sites + TEF constitutive yeast promoter | . . . atacggtcaacgaactataattaactaaac | 457 | 1163 | It's complicated | ||
| BBa_K165034 | Zif268-HIV bs + LexA bs + mCYC promoter | . . . cacaaatacacacactaaattaataactag | 457 | 1431 | It's complicated | ||
| BBa_K165031 | mCYC promoter plus LexA binding sites | . . . cacaaatacacacactaaattaataactag | 403 | 1564 | It's complicated | ||
| BBa_K165030 | mCYC promoter plus Zif268-HIV binding sites | . . . cacaaatacacacactaaattaataactag | 307 | 1723 | It's complicated | ||
| BBa_K165001 | pGAL1+ w/XhoI sites | . . . atactttaacgtcaaggagaaaaaactata | 672 | 2730 | It's complicated | ||
| BBa_K110016 | A-Cell Promoter STE2 (backwards) | . . . accgttaagaaccatatccaagaatcaaaa | 500 | 2256 | It's complicated | ||
| BBa_K110006 | Alpha-Cell Promoter MF(ALPHA)1 | . . . tttcatacacaatataaacgattaaaagaa | 501 | 1773 | It's complicated | ||
| BBa_K110005 | Alpha-Cell Promoter MF(ALPHA)2 | . . . aaattccagtaaattcacatattggagaaa | 500 | 1771 | It's complicated | ||
| BBa_K2601001 | Promoter-PDH3(Saccharomyces cerevisiae) | . . . gtttcgaataaacacacataaacaaacaaa | 500 | 1762 | In stock |
Repressible
All the promoters on this page are yeast promoters that are negatively regulated meaning that increased levels of at least one transcription factor (other than the sigma factor) will decrease the activity of these promoters.
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_K950000 | yeast fet3 promotor | . . . cagtgtaaggaagagtagcaaaaaattaga | 575 | 4852 | In stock | ||
| BBa_K950002 | yeast anb1 promotor | . . . atacacctatttcattcacacactaaaaca | 399 | 5683 | In stock | ||
| BBa_K165000 | MET 25 Promoter | . . . tagatacaattctattacccccatccatac | 387 | 6991 | It's complicated | ||
| BBa_K950003 | yeast suc2 promotor | . . . aaaaagcttttcttttcactaacgtatatg | 699 | 5148 | It's complicated | ||
| BBa_K4706002 | PHO5 Promotor (Phosphate repressed) | . . . caaatagagcaagcaaattcgagattacca | 1084 | ||||
| BBa_I766558 | pFig1 (Inducible) Promoter | . . . aaacaaacaaacaaaaaaaaaaaaaaaaaa | 1000 | 2141 | Not in stock | ||
| BBa_I766214 | pGal1 | . . . atactttaacgtcaaggagaaaaaactata | 1002 | 1190 | Not in stock |
Multiply regulated
This page lists yeast promoters that are Multi-regulated meaning that each promoter is either positively or negatively regulated by multiple transcription factors (other than the sigma factor). For example, a promoter negatively regulated by two repressor proteins forms the basis of a nor gate, the presence of either or both repressors is sufficient to produce a low output from the promoter. These promoters are useful if, for example, you are looking to build logic gates, or if you are looking to build a system where expression of a gene must be dependent on multiple environmental factors.
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_I766200 | pSte2 | . . . accgttaagaaccatatccaagaatcaaaa | 1000 | 1207 | Not in stock | ||
| BBa_K110016 | A-Cell Promoter STE2 (backwards) | . . . accgttaagaaccatatccaagaatcaaaa | 500 | 2256 | It's complicated | ||
| BBa_K165034 | Zif268-HIV bs + LexA bs + mCYC promoter | . . . cacaaatacacacactaaattaataactag | 457 | 1431 | It's complicated | ||
| BBa_K165041 | Zif268-HIV binding sites + TEF constitutive yeast promoter | . . . atacggtcaacgaactataattaactaaac | 457 | 1163 | It's complicated | ||
| BBa_K165043 | Zif268-HIV binding sites + MET25 constitutive yeast promoter | . . . tagatacaattctattacccccatccatac | 441 | 1163 | It's complicated |
Kozak sequences
Yeast RBSs, more often known as Kozak sequences, are designed to be recognized by the yeast ribosome. As a eukaryotic translation signal, the sequence of yeast RBSs are distinct from prokaryotic RBSs. The registry collection of yeast RBSs is currently small, we need your help! Please contribute new yeast RBSs to the registry or provide further information about our existing yeast RBSs.
| Name | Sequence | Description | Relative Strength | Predicted Strength |
|---|---|---|---|---|
| BBa_J63003 | cccgccgccaccatggag | designed yeast Kozak sequence | n/a | |
| BBa_K165002 | cccgccgccaccatggag | Kozak sequence (yeast RBS) | ||
| BBa_K792001 | ggatccacgattaaaagaatg | Kozak sequence from yeast α-factor mating pheromone (MFα1) |
Protein tags and modifiers
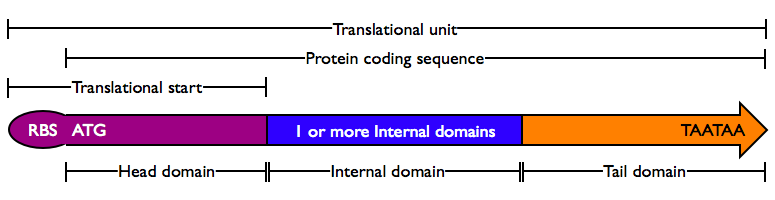
Protein domains encode portions of proteins and can be assembled together to form translational units, a genetic part spanning from translational initiation (the RBS) to translational termination (the stop codon).
There are several different types of protein domains.
- Head Domain: The Head domain consists of the start codon followed immediately by zero or more triplets specifiying an N-terminal tag, such as a protein export tag or lipoprotein binding tag. Head domains should begin with an
ATGstart codon and include codons 2 and 3 of the protein at a minimum. Examples of head domains includeATGstart codonATGstart codon and codons 2-3ATGstart codon and signal sequenceATGstart codon and affinity tag
- Internal Domains: Protein domains consist of a series of codon triplets coding for an amino acid sequence without a start codon or stop codon. Multiple Internal Domains can be fused. Examples of internal domains include
- DNA binding domains
- Dimerization domains
- Kinase domains
- Special Internal Domains: Short Domains with specific function may be separately categorized, but obey the same composition rules as normal internal domains. Examples of special internal domains include
- Linkers
- Cleavage sites
- Inteins
- Tail Domain: The C-terminus of a coding region consists of zero or more triplet codons, followed by a pair of TAA stop codons. In the simplest case, the stop codons terminate the protein with an Stop. More complex Tails may include degradation tags appropriate to the organism (i.e., with different degradation rates). Examples of Tail domain include
TAATAAstop codons- A degradation tag followed by
TAATAAstop codons - An affinity tag followed by
TAATAAstop codon
Unfortunately, the original BioBrick assembly standard, Assembly standard 10, does not support in-frame assembly of protein domains. (Assembly standard 10 creates an 8 bp scar between adjacent parts.) Therefore, it is recommended that you use an alternate approach to assemble protein domains together to make a translational unit. There are several possible approaches to assembling protein domains including direct synthesis (preferred because it creates no scars) as well as various assembly standards. Regardless of which standard you choose, we suggest that the resulting protein coding sequence or translational unit comply with the original BioBrick assembly standard so that your parts can be assembled with most of the parts in the Registry.
Protein coding sequences should be as follows
GAATTC GCGGCCGC T TCTAG [ATG ... TAA TAA] T ACTAGT A GCGGCCG CTGCAG
Note: Although most RBSs are currently specified as separate parts in the Registry, we are now moving to a new design in which the RBS and Head domain are combined into a single part termed a Translational start. The new design has the advantage of encapsulating both ribosome binding and translational initiation within a single part. Our working hypothesis is that the new design will reduce the likelihood of unexpected functional composition problems between the RBS and coding sequence.
| Name | Description | AA sequence | Length |
|---|---|---|---|
| BBa_J63007 | PKI nuclear export sequence; yeast codon optimized | 30 | |
| BBa_J63008 | SV40 nuclear localization sequence from SV40; yeast codon optimized | 21 | |
| BBa_K105013 | cin8 - cell cycle specific degradation tag in yeast | 228 | |
| BBa_K105015 | hsl1 - cell cycle dependent degradation tag in yeast | 618 | |
| BBa_K1486026 | sfGFP + Kanamycin resistance for yeast | 2535 | |
| BBa_K1486027 | R.reniformis luciferase + ADH1 terminator + Kanamycin resistance | 2681 | |
| BBa_K1486028 | Yeast optimized sfGFP N-terminus (1-214) | 642 | |
| BBa_K1486033 | R.reniformis luciferase + ADH1 terminator + CaUra3 | 2523 | |
| BBa_K1486034 | Yeast optimized superfolder GFP C-terminus (215-238) | 75 | |
| BBa_K1486035 | Yeast Optimized sfGFP-C + ADH1 terminator + CaUra3 Cassette | 1662 | |
| BBa_K1486036 | rLucC + ADH1 terminator + CaUra3 cassette | 2193 | |
| BBa_K1680004 | SV40 NLS | 24 | |
| BBa_K416000 | Aga2 CDS Responsible for Yeast Surface Display | 261 | |
| BBa_K416003 | Yeast Secretion Tag | 114 | |
| BBa_K4706010 | SV40 NLS codon optimised S. cerevisae | 51 | |
| BBa_K5466002 | AGA2P Signal Peptide | 60 | |
| BBa_K5466006 | SUC2 Signal Peptide. Saccharomyces cerevisiae codon optimized | 57 | |
| BBa_K792002 | Secretion tag from yeast α-factor mating pheromone (MFα1) | 54 |
Protein coding sequences
Protein coding sequences are DNA sequences that are transcribed into mRNA and in which the corresponding mRNA molecules are translated into a polypeptide chain. Every three nucleotides, termed a codon, in a protein coding sequence encodes 1 amino acid in the polypeptide chain. In some cases, different chassis may either map a given codon to a different sequence or may use different codons more or less frequently. Therefore some protein coding sequences may be optimized for use in a particular chassis.
In the Registry, protein coding sequences begin with a start codon (usually ATG) and end with a stop codon (usually with a double stop codon TAA TAA). Protein coding sequences are often abbreviated with the acronym CDS.
Although protein coding sequences are often considered to be basic parts, in fact proteins coding sequences can themselves be composed of one or more regions, called protein domains. Thus, a protein coding sequence could either be entered as a basic part or as a composite part of two or more protein domains.
- The N-terminal domain of a protein coding sequence is special in a number of ways. First, it always contains a start codon, spaced at an appropriate distance from a ribosomal binding site. Second, many coding regions have special features at the N terminus, such as protein export tags and lipoprotein cleavage and attachment tags. These occur at the beginning of a coding region, and therefore are termed Head domains.
- A protein domain is a sequence of amino acids which fold relatively independently and which are evolutionarily shuffled as a unit among different protein coding regions. The DNA sequence of such domains must maintain in-frame translation, and thus is a multiple of three bases. Since these protein domains are within a protein coding sequence, they are called Internal domains. Certain Internal domains have particular functions in protein cleavage or splicing and are termed Special Internal domains.
- Similarly, the C-terminal domain of a protein is special, containing at least a stop codon. Other special features, such as degradation tags, are also required to be at the extreme C-terminus. Again, these domains cannot function when internal to a coding region, and are termed Tail domains.
For more details on protein domains including how to assemble protein domains into protein coding sequences, please see Protein domains.
Protein coding sequences should be as follows
GAATTC GCGGCCGC T TCTAG [ATG ... TAA TAA] T ACTAGT A GCGGCCG CTGCAG
| Name | Protein | Description | Tag | Direction | UniProt | KEGG | Length | Status |
|---|---|---|---|---|---|---|---|---|
| BBa_E2050 | mOrange | derivative of mRFP1, yeast-optimized | None | 769 | In stock | |||
| BBa_K106001 | Sir4, Aar1 AD part | 4077 | It's complicated | |||||
| BBa_K106002 | Sir2, Aar1 AB part | 1686 | It's complicated | |||||
| BBa_K106003 | Sir2, Aar1 BD part | 1686 | It's complicated | |||||
| BBa_K106011 | Sas2 | Sas2 histone acetyltransferase, Aar1 AB part | 1017 | It's complicated | ||||
| BBa_K106012 | Sas2 | Sas2 histone acetyltransferase, Aar1 BD part | 1014 | It's complicated | ||||
| BBa_K106013 | Esa1 | Esa1 histone acetyltransferase, Aar1 BD part | 1338 | It's complicated | ||||
| BBa_K1314017 | Mid1 (S. cerevisae Codon Opt) | 1680 | It's complicated | |||||
| BBa_K1462000 | CoxVI | 120 | Not in stock | |||||
| BBa_K1462010 | coxIV | 75 | Not in stock | |||||
| BBa_K1462020 | Erg10 | 1197 | Not in stock | |||||
| BBa_K1462030 | Hbd | 849 | Not in stock | |||||
| BBa_K1462050 | Ccr | 1344 | Not in stock | |||||
| BBa_K1462060 | AdhE2 | 2577 | Not in stock | |||||
| BBa_K1592000 | LIP2 prepro(signal peptide) | 99 | In stock | |||||
| BBa_K1611001 | MATalpha-IFNgamma | 735 | It's complicated | |||||
| BBa_K1611002 | OVA1 | 66 | In stock | |||||
| BBa_K1611004 | DEC-205 | 801 | In stock | |||||
| BBa_K165005 | Venus YFP, yeast optimized for fusion | Forward | 744 | In stock | ||||
| BBa_K1680006 | Fluorescent protein dronpa | 669 | In stock | |||||
| BBa_K1680007 | Cre recombinase | 1029 | In stock | |||||
| BBa_K1680008 | Δ1-19 Cre recombinase | 972 | In stock | |||||
| BBa_K1680009 | NanoLuc Luciferase | 516 | In stock | |||||
| BBa_K1680010 | Firefly Luciferase | 1653 | In stock | |||||
| BBa_K1680011 | HCV protease with linkers | 648 | In stock | |||||
| BBa_K1680012 | Cleavage site for HCV protease | 36 | In stock | |||||
| BBa_K1680013 | NDegron | 165 | In stock | |||||
| BBa_K1680017 | Cre-Dronpa Fusion | 1728 | It's complicated | |||||
| BBa_K1680018 | Cre-Dronpa fusion | 1746 | It's complicated | |||||
| BBa_K1680019 | Cre-dronpa fusion | 1764 | It's complicated | |||||
| BBa_K1680020 | Cre-dronpa fusion | 1782 | It's complicated | |||||
| BBa_K1680021 | Dronpa caged Cre with NLS | 2433 | It's complicated | |||||
| BBa_K1680022 | Dronpa caged Cre with NLS | 2451 | It's complicated | |||||
| BBa_K1680023 | Dronpa caged Cre with NLS | 2469 | It's complicated | |||||
| BBa_K1680024 | Dronpa caged Cre with NLS | 2487 | It's complicated | |||||
| BBa_K1728021 | Chemokine receptor 1 (CXCR1) | 1053 | Not in stock | |||||
| BBa_K1907000 | Venus Yellow Fluorescent Protein | 717 | It's complicated | |||||
| BBa_K1907001 | Microcystinase (MlrA) for S. cerevisiae | 1038 | It's complicated | |||||
| BBa_K1907006 | Microcystinase (Mlra) + mating factor alpha tag, for S. cerevisiae | 1305 | Not in stock | |||||
| BBa_K1908001 | Coding sequence for Homo sapien Cripto-1 membrane protein. | 767 | It's complicated | |||||
| BBa_K211002 | RI7-odr10 chimeric GPCR | FLAG | 1062 | It's complicated | ||||
| BBa_K2225000 | AtNHXS1 for S. cerevisiae (Na+/H+ antiporter) | 729 | It's complicated | |||||
| BBa_K2225002 | AtSultr1;2 for S.cerevisiae (high-affinity sulfate transporter ) | 1962 | It's complicated | |||||
| BBa_K2247002 | HXT2 glucose transpoter from S. cerevisiae | 1623 | It's complicated | |||||
| BBa_K2247003 | GFP tagged HXT2 glucose transpoter from S. cerevisiae | 2349 | It's complicated | |||||
| BBa_K2247004 | HXT5 glucose transpoter from S. cerevisiae | 1776 | It's complicated | |||||
| BBa_K2247005 | GFP tagged HXT5 glucose transpoter from S. cerevisiae | 2502 | It's complicated | |||||
| BBa_K2256000 | sod2 | sod2 (Na+/H+antiporter) from Schizosaccharomyces pombe | 657 | It's complicated | ||||
| BBa_K2483007 | Ddx4-YFP | Ddx4-YFP | 1710 | It's complicated | ||||
| BBa_K2583006 | GPA1 | Coding sequence of GPA1 in yeast genome | 1419 | It's complicated | ||||
| BBa_K2601002 | SUMO (Small Ubiquitin-like Modifier) | 273 | It's complicated | |||||
| BBa_K2601003 | SIM (SUMO-Interaction Motif) | 69 | It's complicated | |||||
| BBa_K2601004 | HOTag3 (Homo-Oligomeric Tag3) | 90 | It's complicated | |||||
| BBa_K2601005 | HOTag6 (Homo-Oligomeric Tag6) | 99 | It's complicated | |||||
| BBa_K2637001 | KaiA Protein of Circadian Rhythm | Forward | 858 | It's complicated | ||||
| BBa_K2637002 | KaiB Protein of Circadian Rhythm | Forward | 312 | It's complicated | ||||
| BBa_K2637003 | KaiC Protein of Circadian Rhythm | Forward | 1563 | It's complicated | ||||
| BBa_K2637004 | SasA Protein of Circadian Rhythm | Forward | 1167 | It's complicated | ||||
| BBa_K2637005 | CikA Protein of Circadian Rhythm | Forward | 2268 | It's complicated | ||||
| BBa_K2637006 | RpaA Protein of Circadian Rhythm | Forward | 753 | It's complicated | ||||
| BBa_K2637007 | AD-Activation Domain of Gal4 transcription factors | Forward | 402 | It's complicated | ||||
| BBa_K2637008 | BD-DNA-binding Domain of Gal4 transcription factors | Forward | 457 | It's complicated | ||||
| BBa_K2637010 | Nanoluc+PEST | Forward | 642 | It's complicated | ||||
| BBa_K2835005 | His-tagged fungal laccase with start codon (ATG) | 6X His | 1521 | It's complicated | ||||
| BBa_K284000 | Lactate Permease from Kluyveromyces lactis | 1873 | Not in stock | |||||
| BBa_K3559000 | BioBrick_BASIC_DAHP Synthase (Aro4) (K229L) | 1163 | Not in stock | |||||
| BBa_K3629008 | Modified Trichoderma reesei EGI with 6X His tag | 1158 | Not in stock | |||||
| BBa_K3629009 | Trichoderma reesei EGII with FLAG tag | 1242 | Not in stock | |||||
| BBa_K3629010 | Neocallimastix patriciarum BGS with 6X His tag | 2313 | Not in stock | |||||
| BBa_K3629011 | Nourseothricin resistance gene | 573 | Not in stock | |||||
| BBa_K3629020 | Yarrowia lipolytica TRP2 Gene | 1524 | Not in stock | |||||
| BBa_K3629021 | mCherry codon optimized for Yarrowia lipolytica | 711 | Not in stock | |||||
| BBa_K3629022 | mCitrine codon optimized for Yarrowia lipolytica | 720 | Not in stock | |||||
| BBa_K3772005 | valencene synthase | VS, valencene synthase coding sequence | 1770 | |||||
| BBa_K3819007 | Cellulase | EG1(Trichoderma sp. SSL endoglucanase I) | 1383 | |||||
| BBa_K3819015 | CtDockerin domain | CtDockerin (TypeⅠ from Clostridium thermocellum) | 207 | |||||
| BBa_K3819062 | XynB | XynB (Xylanase, Aspergillus niger) | 675 | |||||
| BBa_K394000 | Resistance Gene for G418 in Yeast | 813 | It's complicated | |||||
| BBa_K394003 | Sex-Lethal from Drosophila melanogaster | 1062 | Not in stock | |||||
| BBa_K4035001 | CUP1 fused to Aga2 and tagged with a V5 epitope | Forward | 612 | Not in stock | ||||
| BBa_K4035002 | Dimerization of the copper metallothionein 1 : CUP1-(GGGGS)3-CUP1 | Forward | 405 | Not in stock | ||||
| BBa_K4035003 | Dimerization of the copper metallothionein 1 : CUP1-(GGGGS)4-CUP1 | Forward | 420 | |||||
| BBa_K4035004 | Dimerization of the copper metallothionein 1 : CUP1-(AP)7-CUP1 | Forward | 402 | |||||
| BBa_K4035005 | Dimerization of the copper metallothionein 1 : CUP1-(EAAAK)3-CUP1 | Forward | 405 | |||||
| BBa_K4035006 | Dimerization of the copper metallothionein 1 : CUP1-(EAAAK)4-CUP1 | Forward | 420 | |||||
| BBa_K4035007 | Dimerization of the copper metallothionein 1 : CUP1-GGGGSEAAAKGGGGS-CUP1 | Forward | 405 | |||||
| BBa_K4035008 | Dimerization of the copper metallothionein 1 : CUP1-GGGGS(EAAAK)2GGGGS-CUP1 | Forward | 420 | |||||
| BBa_K4298002 | enCas9-PolI5M | 7698 | ||||||
| BBa_K4298005 | TM8.3 gene | 143 | ||||||
| BBa_K4298006 | TM8.3 with α-factor secretion signal | 930 | ||||||
| BBa_K4298013 | yEmCitrine | 925 | ||||||
| BBa_K4706005 | Red fluorescent GFP-like protein | meffRed/meffRFP codon optimised for S. cerevisiae | none | forward | A8CLN4_MONEF | 702 | ||
| BBa_K4706006 | 2A self cleaving peptide S. cerevisae | E2A self cleaving peptide | none | forward | 60 | |||
| BBa_K4706007 | BCL-2-associated athanogene 6 | AtBAG6 induced S. cerevisae cell death | none | forward | At2g46240 | 405 | ||
| BBa_K4706008 | Green fluorescent GFP-like protein | eechGFP1 codon optimised S.cerevisae | none | forward | A8CLQ1_9CNID | 687 | ||
| BBa_K4706009 | HTH-type transcriptional regulator SrpR | SrpR codon optimised S. cerevisae | none | forward | Q9R9T9 � SRPR_PSEPU | 639 | ||
| BBa_K4795001 | Encodes endoglucanase | 666 | ||||||
| BBa_K4795002 | Encodes exoglucanase | 909 | ||||||
| BBa_K4795003 | Coding for β-glucosidase | 1341 | ||||||
| BBa_K4795013 | Code for xylose isomerase | 1311 | ||||||
| BBa_K4795017 | Regulation of xylose isomerase expression | 2814 | ||||||
| BBa_K4795019 | Regulation of xylose isomerase expression | 1961 | ||||||
| BBa_K4795020 | Regulation of xylose isomerase expression | 1661 | ||||||
| BBa_K5146000 | Cellulose binding domain attached to a mating factor secretion tag | BT1_preOst1_proalpha_MF_signal | 813 | |||||
| BBa_K5166000 | Nickel-Binding Peptide 1 | 36 | ||||||
| BBa_K5166001 | Nickel-Binding Peptide 2 | 36 | ||||||
| BBa_K5166002 | Nickel-Binding Peptide 3 | 36 | ||||||
| BBa_K5166004 | Cobalt-Binding Peptide 1 | 36 | ||||||
| BBa_K5166005 | Cobalt-Binding Peptide 2 | 36 | ||||||
| BBa_K5166006 | Lithium-Binding Peptide 3 | 45 | ||||||
| BBa_K5166007 | MntR | 423 | ||||||
| BBa_K5166008 | TssS | 144 | ||||||
| BBa_K5166009 | BH2807 | 417 | ||||||
| BBa_K5466010 | LexA-VP16 | 897 | ||||||
| BBa_K5466017 | Nb28-S102D Aga2P | 681 | ||||||
| BBa_K5466018 | antiAFB1-scFv1 NubG | 1410 | ||||||
| BBa_K5466019 | antiAFB1-scFv2 Cub | 2322 | ||||||
| BBa_K5466031 | AntiAFB1-scFv1 EpoR Split-N-mCerulean | 1809 | ||||||
| BBa_K5466032 | AntiAFB1-scFv2 EpoR Split-C-mCerulean | 1542 | ||||||
| BBa_K775000 | RI7-GPR109A Niacin chimeric GPCR | 1414 | In stock | |||||
| BBa_K775003 | RI7-odr10 (diacetyl) chimeric GPCR with promoter and terminator | 1444 | In stock | |||||
| BBa_K801039 | SV40NLS-GAL4AD-linker-PIF3 (part for GAL4/LexA based light-switchable promoter system) | 804 | In stock | |||||
| BBa_K801040 | SV40NLS-PhyB-linker-Gal4DBD (part for GAL4 based light switchable promoter system) | 3294 | In stock | |||||
| BBa_K801060 | (+)-Limonene synthase 1 with Strep-tag and yeast consensus sequence. | 1708 | In stock | |||||
| BBa_K801061 | (+)-Limonene synthase 1 coding region | 1665 | In stock | |||||
| BBa_K801070 | xanthosine methyltransferase CaXMT1-strep | 1158 | In stock | |||||
| BBa_K801071 | 7-methylxanthine N-methyltransferase CaMXMT1-strep | 1176 | In stock | |||||
| BBa_K801072 | 3,7-dimethylxanthine N-methyltransferase CaDXMT1-strep | 1194 | In stock | |||||
| BBa_K801080 | Prepro-Thaumatin, yeast codon optimized | 708 | In stock | |||||
| BBa_K801090 | phenylalanine ammonia lyase (PAL) + yeast consensus sequence | 2154 | In stock | |||||
| BBa_K801091 | phenylalanine ammonia lyase (PAL) coding region | 2148 | In stock | |||||
| BBa_K801092 | 4-coumarate - coenzym A ligase (4CL) + yeast consensus sequence | 1692 | In stock | |||||
| BBa_K801093 | 4-coumarate--coenzyme A ligase (4CL) coding region | 1686 | In stock | |||||
| BBa_K801094 | Naringenin - chalcone synthase (CHS) + yeast consensus sequence | 1176 | In stock | |||||
| BBa_K801095 | Naringenin - chalcone synthase (CHS) coding region | 1170 | In stock | |||||
| BBa_K801096 | Aromatic prenyltransferase (APT) coding region | 1233 | In stock | |||||
| BBa_K801097 | Chalcone O-methyltransferase (OMT1) + yeast consensus sequence | 1062 | In stock | |||||
| BBa_K801098 | O-methyltransferase 1 (OMT1) coding region | 1056 | In stock | |||||
| BBa_K809401 | REX coding sequence | 777 | Not in stock | |||||
| BBa_K950001 | yeast rox1 | 1225 | In stock | |||||
| BBa_K950009 | yeast mig1 | 1515 | In stock | |||||
| BBa_Y00029 | S. pombe homolog of S.cerevisiae SGF29, recoded for expression in S.c. | 794 | It's complicated | |||||
| BBa_Y00073 | S. pombe gene SPCC126.04c, coded for S. cerevisiae expression | 1094 | It's complicated |
Translational units
Translational units begin with the RBS, the site of ribosome binding and translational initiation, and end with a stop codon, the site of translational termination. Every translational unit in the Registry consists of at least three parts, a Translational start, one or more Internal Domains including Special Internal Domains, and a Tail Domain. Thus translational units can, in some sense, be thought of as a composite part made up of three or more parts. Protein coding sequences, in contrast, begin with a start codon and end with a stop codon.
For more information on protein domains, see protein domains. Unfortunately, the original BioBrick assembly standard, Assembly standard 10, does not support in-frame assembly of protein domains. (Assembly standard 10 creates an 8 bp scar between adjacent parts.) Therefore, it is recommended that you use an alternate approach to assemble protein domains together to make a translational unit. There are several possible approaches to assembling protein domains including direct synthesis (preferred because it creates no scars) as well as various assembly standards. Regardless of which standard you choose, we suggest that the resulting translational unit comply with the original BioBrick assembly standard so that your parts can be assembled with most of the parts in the Registry.
Translational units should be as follows
GAATTC GCGGCCGC T TCTAGA G [RBS] [ATG ... TAA TAA] T ACTAGT A GCGGCCG CTGCAG
Although most RBSs are currently specified as separate parts in the Registry, we are now moving to a new design in which the RBS and Head domain are combined into a single part termed a Translational start. The new design has the advantage of encapsulating both ribosome binding and translational initiation within a single part. Our working hypothesis is that the new design will reduce the likelihood of unexpected functional composition problems between the RBS and coding sequence.
| Name | Protein | Description | Tag | Direction | UniProt | KEGG | Length | Status |
|---|---|---|---|---|---|---|---|---|
| BBa_K165057 | Kozak + mCherry | 761 | It's complicated | |||||
| BBa_K165058 | Kozak + YFP | 770 | It's complicated | |||||
| BBa_K165059 | Kozak + CFPx2 | 1520 | It's complicated | |||||
| BBa_K976009 | TDH3 Promoter + Yeast Kozak + FGFR-1/FRS2 Complex +TEF1 Terminator | 5224 | Not in stock |
Terminators
Here are all the yeast terminators available. As you can see, there are only a couple available, so please design, construct, and characterize new ones and submit them to the Registry!
| Name | Description | Direction | Efficiency Fwd. Rev. | Chassis | Length | |
|---|---|---|---|---|---|---|
| BBa_J63002 | ADH1 terminator from S. cerevisiae | Forward | 225 | |||
| BBa_K110012 | STE2 terminator | Forward | 123 | |||
| BBa_K1462070 | cyc1 | 250 | ||||
| BBa_K1486025 | ADH1 Terminator | Forward | Saccharomyces Cerevisiae | 188 | ||
| BBa_K2314608 | Tmini is a very short terminator in yeast. It's only 68bp in length but has a good performance. | 68 | ||||
| BBa_K2637012 | ADH1 Terminator | S. cerevisiae | 335 | |||
| BBa_K2637014 | TEF1 Terminator | S. cerevisiae | 476 | |||
| BBa_K2637016 | PGK1 Terminator | S. cerevisiae | 285 | |||
| BBa_K2637017 | CYC1 terminator | 261 | ||||
| BBa_K3768004 | CPS1 Terminator for S. cerevisiae | 191 | ||||
| BBa_K392003 | yeast ADH1 terminator | 129 | ||||
| BBa_K4947020 | Yeast terminator with I-SceI restriction site | 50 | ||||
| BBa_K801011 | TEF1 yeast terminator | 507 | ||||
| BBa_K801012 | ADH1 yeast terminator | 349 | ||||
| BBa_Y1015 | CycE1 | 252 | ||||
| Caroline Ajo-Franklin developed the yeast terminator BBa_J63002 as a post-doc in Pam Silver's lab. |
References
- [http://www.yeastgenome.org/ Yeast Genome Database]
- [http://mips.gsf.de/genre/proj/yeast/ MIPS Saccharomyces cerevisiae genome database]
- [http://www.yeastgenome.org/VL-yeast.html Virtual Library on yeast]