Part:BBa_K4035001
CUP1 fused to Aga2 and tagged with a V5 epitope
This protein is made of the copper metallothionein CUP1 (BBa_M45090) fused to the A-agglutinin-binding subunit Aga2 (BBa_K416000) at its N-terminal and to the V5 tag (BBa_T2020) at its C-terminal.
Usage and Biology
Copper metallothionein CUP1 (BBa_M45090) is a protein responsible for copper binding protein in the yeast Saccharomyces cerevisiae. Normally expressed intracellularly, CUP1 was fused to the A-agglutinin-binding subunit Aga2 (BBa_K416000) that attaches to the yeast cell wall through disulfide bonds to Aga1p. This leads to the presence of CUP1 on the outer membrane of the cell. The goal behind it was to improve the endogenous bio-accumulation property of wild type yeast by allowing it to bind more copper ions on its external surface.
This fusion protein also contains a V5 tag (BBa_T2020) in order to check its expression by Western Blot and Immunocytochemistry. The expression is under the control of the Gal1 promoter (BBa_J63006), so that the protein is expressed only in the presence of galactose.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 319
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 376
Illegal PstI site found at 319 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 562
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 319
- 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 319
- 1000COMPATIBLE WITH RFC[1000]
Characterization
Expression of the protein in S.cerevisiae
Western Blot Analysis
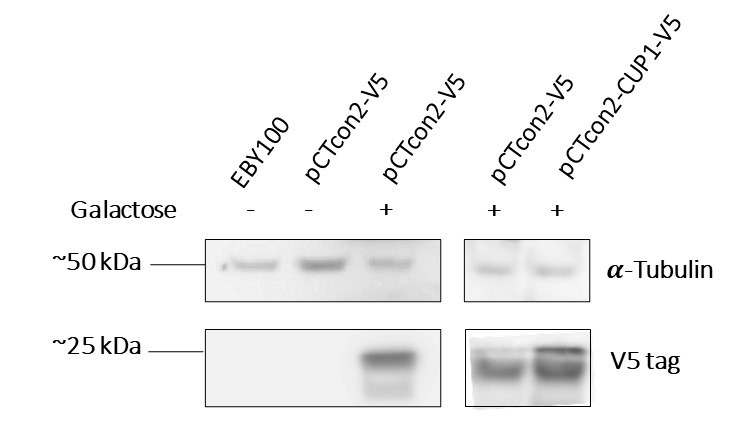
In order to characterize the protein expression, two experiments were performed, the first being a Western Blot analysis. After having transformed the EBY100 yeast with our newly formed plasmid pCTcon2-CUP1-V5 we tested its protein expression. For control we also tested the wild type yeast (untransformed) as well as a transformed yeast with the plasmid backbone (without insert) in two different media, one containing galactose and allowing expression and the other, lacking galactose and thus blocking expression of the protein. As the plasmid contains a Gal1 promoter, the system can only be expressed in the presence of galactose.
Figure 1a : The first two lanes, EBY100 and pCTcon2-V5 without galactose which are, respectively, wild type yeast and uninduced transformed yeast with the backbone plasmid, serve as negative controls, and show no presence of the V5 tag, as we expected. The third and fourth lanes, pCTcon2-V5 + galactose, are the induced transformed yeast with the backbone plasmid and show expression of the V5 tag which proves that our system is expressed in the transformed yeast when induced with galactose. The last lane is our transformed yeast with the plasmid pCTcon2-CUP1-V5 and also shows expression of the system.
We can see that we have two bands on the gel, one is of approximately 25 kDa which is the size of the fusion protein Aga2-CUP1-V5 and the other is slightly smaller. The smaller band could be a truncated version of the protein since we identified a second in-frame start codon in the DNA sequence. Unfortunately the truncated version seems to be the most expressed one, corresponding to the signal intensity.
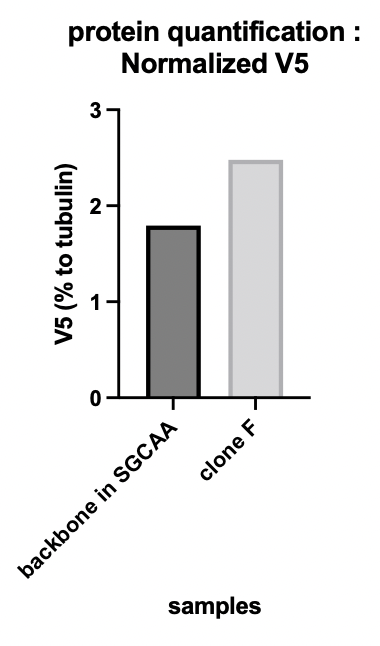
The alpha-tubulin signal allowed us to quantify the protein expression. Figure 1b shows the V5 tag relative expression quantification. The signal of V5 has been normalized by the tubulin signal. Clone F means the transformed yeast clone and SGCAA is the yeast selection medium containing galactose.
The presence of our membrane protein Aga2-CUP1-V5 in our yeast transformants is thus shown.
Immunocytochemistry Assay
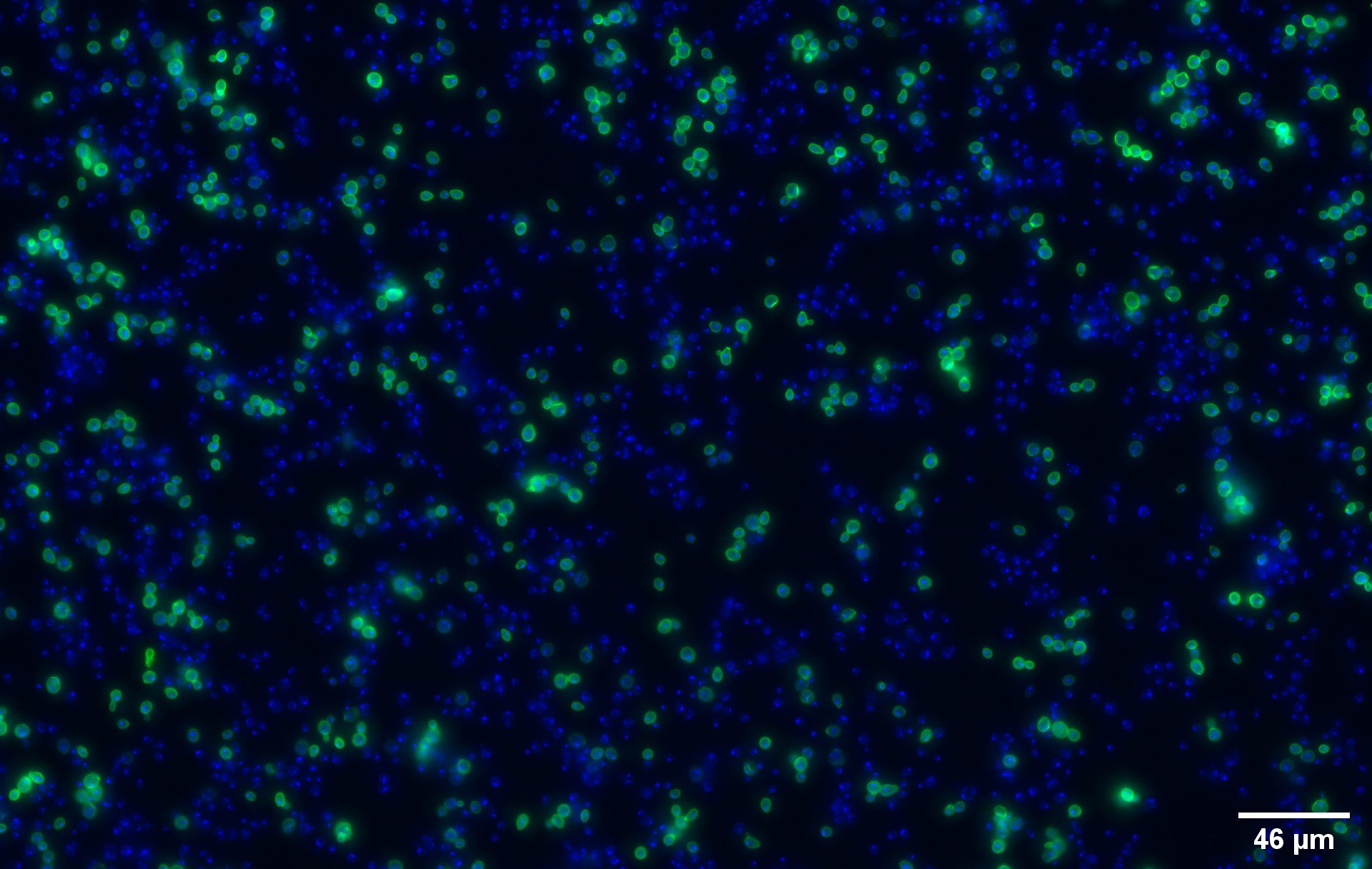
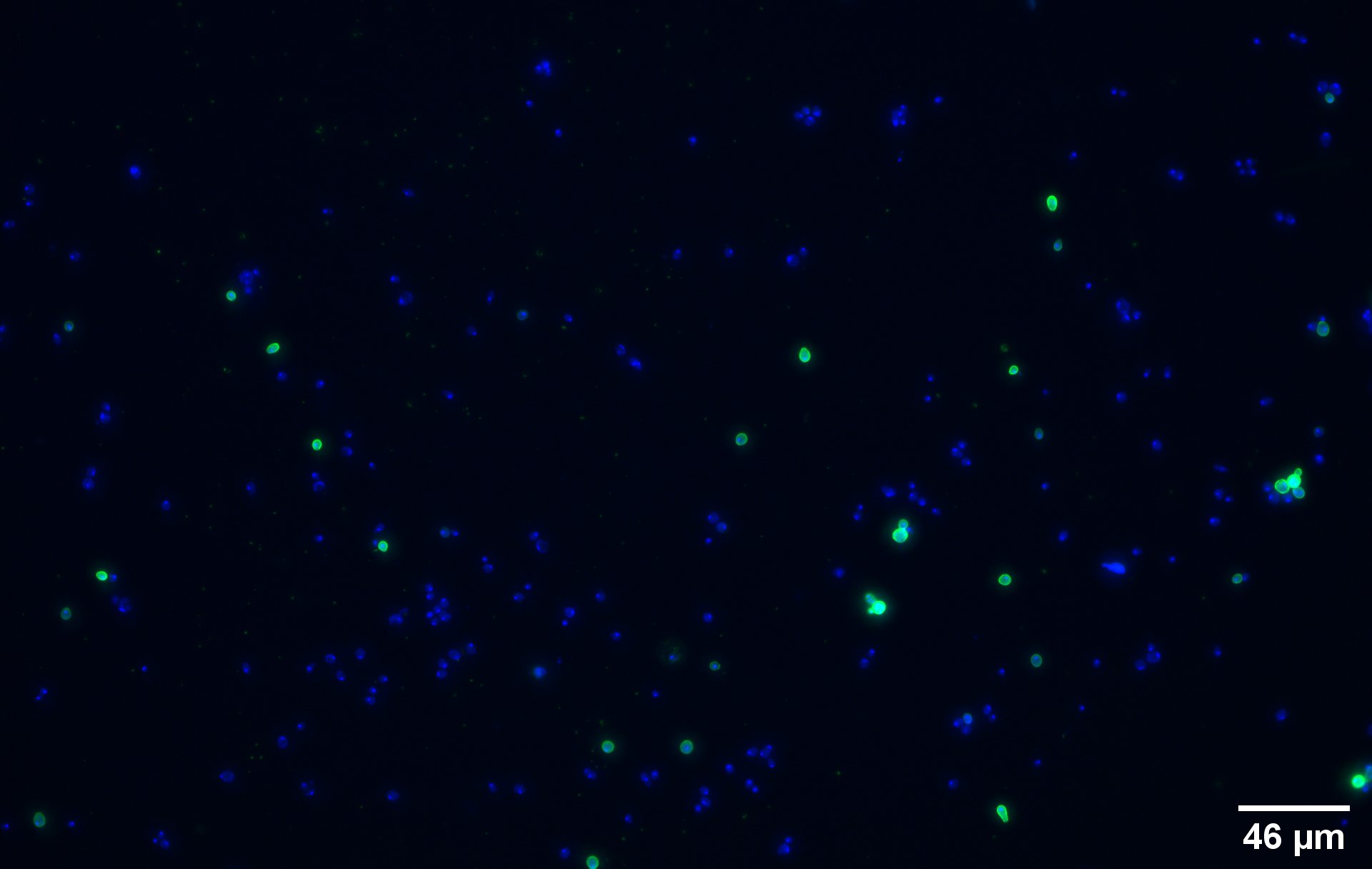
To show that the fusion protein is expressed at the membrane of the cell we performed an Immunocytochemistry Assay. The cells are incubated with a primary mouse anti-V5 antibody as well as with a secondary goat anti-mouse couple with Alexa Fluor 488. The same strains for control have been used, namely, plasmid backbone induced (Figure 2c) and wild type yeast (Figure 2d).
The nuclei have been stained with DAPI and are depicted in blue on the figures. The green disks are representing the transformed yeast cells expressing CUP1 at their surface (Figure 2a).
Due to the more intense circle we can clearly see that our system is expressed on the membrane of the protein (better seen on Figure 2b).
We can also remark that not all the cells are expressing the system. This is because the expression is not 100% efficient, concordant to the published expression system (1).
On the negative control, the little green signal we see is background noise or nonspecific antibody binding.
Growth and Survival Characterization
Growth curves of the different yeast strains
To check if our expression system would affect the growth of our micro-organism, we performed multiple growth curves with different parameters and compared the results with the wild type strain as well as the yeast transformed with the plasmid backbone in SDCAA (uninduced expression) and in SGCAA (induced expression). We tested the growth of the different strains in medium containing copper in order to see if copper would affect their growth. The results are shown on the figures below.
The transformed yeasts grew slower in selection media SGCAA and SDCAA than the wild type yeast in YPD media which is a complete yeast medium composed of yeast extract peptone dextrose. Addition of copper in the media did not have a high impact on the yeast growth, proving that the microorganisms were quite resistant.
However, we can observe that the expression of the system did affect the growth of the yeast (Figure 3c and 3d).
Copper Absorption
To finally test our system we designed an experiment to measure how much copper our transformed yeast could absorb. We cultured different yeast strains in a medium containing a defined initial concentration of copper and collected samples at different time points to measure their copper concentration. The detailed protocol can be found [ here].
Wild type yeasts (EBY100) absorbed copper well. However, we were not able to make our engineered yeast with the CUP1 monomer work better (Figure 4a).
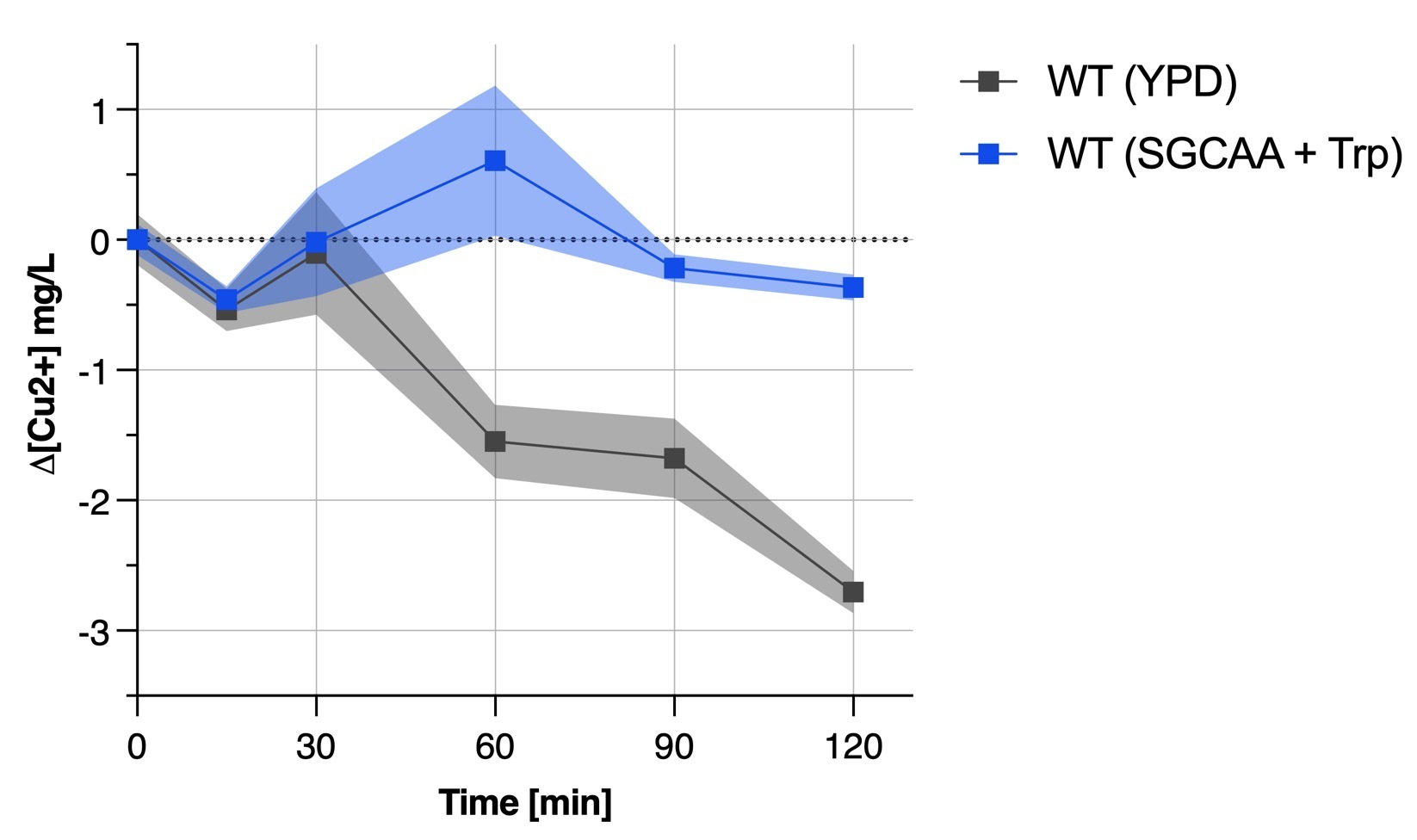
We found out that the yeast medium was an important factor when culturing yeast for bioremediation since our transformant did not show any results when in SGCAA whereas was acting like the wild type yeast when in YPD rich medium. This is also proved by Figure 4b depicting the unability of the wild type yeast in SGCAA + tryptophan to remove copper from solution.
//cds/membrane/extracellular
//chassis/eukaryote/yeast
| biology | CUP1 |
| chassis | S.cerevisiae |
| control | pCTcon2V5, WT EBY100 |
| direction | Forward |
| function | copper binding protein |