Difference between revisions of "Biosafety"
(→All biosafety part) |
|||
| (31 intermediate revisions by 5 users not shown) | |||
| Line 6: | Line 6: | ||
#assembly_plasmid_table td {background-color:#eeeeee; padding: 5px; font-size: 12px;} | #assembly_plasmid_table td {background-color:#eeeeee; padding: 5px; font-size: 12px;} | ||
#assembly_plasmid_table th {background-color: #aaaaaa;padding: 5px; font-size:14px;} | #assembly_plasmid_table th {background-color: #aaaaaa;padding: 5px; font-size:14px;} | ||
| + | ul { | ||
| + | list-style-type:disc; | ||
| + | list-style-image:none; | ||
| + | } | ||
| + | |||
| + | #tableboxed { | ||
| + | border-color: #559DD5; | ||
| + | border-radius: 20px 20px 20px 20px; | ||
| + | border-style: solid; | ||
| + | border-width: 7px; | ||
| + | margin-left: auto; | ||
| + | margin-right: auto; | ||
| + | } | ||
| + | |||
| + | |||
| + | #tableboxed td { | ||
| + | padding: 20px; | ||
| + | border:none; | ||
| + | } | ||
| + | |||
</style> | </style> | ||
</html> | </html> | ||
| Line 12: | Line 32: | ||
* Quick links of pubmed research : | * Quick links of pubmed research : | ||
| − | #[http://www.ncbi.nlm.nih.gov/pubmed?term=biosafety%20gmo | + | #[http://www.ncbi.nlm.nih.gov/pubmed?term=biosafety%20gmo Biosafety and GMO's] |
| − | #[http://www.ncbi.nlm.nih.gov/pubmed?term=biosafety%20synthetic%20biology | + | #[http://www.ncbi.nlm.nih.gov/pubmed?term=biosafety%20synthetic%20biology Biosafety in Synthetic Biology] |
| − | #[http://www.ncbi.nlm.nih.gov/pubmed?term=issue%20synthetic%20biology | + | #[http://www.ncbi.nlm.nih.gov/pubmed?term=issue%20synthetic%20biology Issues of Synthetic Biology] |
| − | We encourage you to do this kind of research as part of your bibliography for whatever | + | We encourage you to do this kind of research as part of your bibliography for whatever project you are working on. |
| − | * Here is the link an openwetware page that | + | * Here is the link to an openwetware page that should be improved by the community. This report exists to improve biosafety practices in synthetic biology, raise good questions, and encourage collaboration in exploring the issue. |
| − | ->[http://openwetware.org/wiki/How_safe_is_safe_enough:_towards_best_pratices_of_synthetic_biology How safe is | + | -> [http://openwetware.org/wiki/How_safe_is_safe_enough:_towards_best_pratices_of_synthetic_biology How safe is Safe Enough?] |
| − | * | + | * Link to the biosafety recommendation page of iGEM: |
[https://igem.org/safety Safety page] | [https://igem.org/safety Safety page] | ||
| − | It includes book, | + | It includes book, papers and other useful references. |
| + | |||
| + | ==Recommendation from Human Practice Teams== | ||
| + | |||
| + | * Report of Sara Aguiton : [http://2009.igem.org/Team:Paris/Ethics_ethicalreport#top report] | ||
| + | * Report of Claire Mayer : [http://2012.igem.org/Team:Paris_Bettencourt/Human_Practice/Report report] | ||
| + | |||
| + | <table id="tableboxed"> | ||
| + | <tr> | ||
| + | <td> | ||
| + | ''' Conclusion of these reports : ''' | ||
| + | * daily and local efforts : An ethical reflection on practices, discourses and social interactions. | ||
| + | * A responsible position regarding what scientific paradigms, through concepts, perceptions and values, are emerging with this field. | ||
| + | *The need to raise awareness of synthetic biology in the population so people can decide in the most enlightened way possible if they want of this new technology and its applications. | ||
| + | * The need of a discussion between society’s different decision makers to set goals and a definition of what they would consider benefits and acceptable risks. | ||
| + | * Zero risk is impossible to achieve as no containment system can be 100% safe (bacteria can always escape). | ||
| + | * There is a lack of quantitative data evaluating the probability of failure of any synthetic biology engineered system, in particular containment systems. | ||
| + | * There is a lack of quantitative data evaluating the risk of HGT assuming containment systems failed. | ||
| + | * The compiling of the wiki screen shows that no containment systems created in iGEM is robust: they lack the above quantification and are mostly one mutation away from failure. We call for major effort of the iGEM community to quantify available containment systems and search for new solutions. | ||
| + | * The need for an INDEPENDENT cohort of scientists to test experimentally any application of synthetic biology that requires release into the environment. | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | ==Proposals to improve this page (Feel free to add ideas)== | ||
| + | |||
| + | # Add a biosafety grade, that could quantify the degree of robustness. For instance, in semantic containment with amber mutation, we score the robustness with the number of amber mutations. | ||
| + | |||
| + | ==Example of biosafety systems == | ||
| + | |||
| + | Here are reported biosafety systems that teams created. The risk and hazards of these systems are assessed through fault tree analysis, a top-down approach used in engineering field. | ||
| + | |||
| + | <table id=""> | ||
| + | <tr> | ||
| + | <th>Team</th> | ||
| + | <th>Fault Tree Analysis</th> | ||
| + | <th>Guesstimation</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Paris Bettencourt 2012</td> | ||
| + | <td>[[Image:FTA_PB12.png|thumb|350px]]</td> | ||
| + | <td><table style="border-spacing:0;"> | ||
| + | <tr> | ||
| + | <td style="border:0.035cm solid #000000;padding:0.176cm;"> <b>Notation</b> | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> <b>Failure component</b> | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> <b>Failure mode</b> | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> <b>Consequence</b> | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> <b>Probability</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
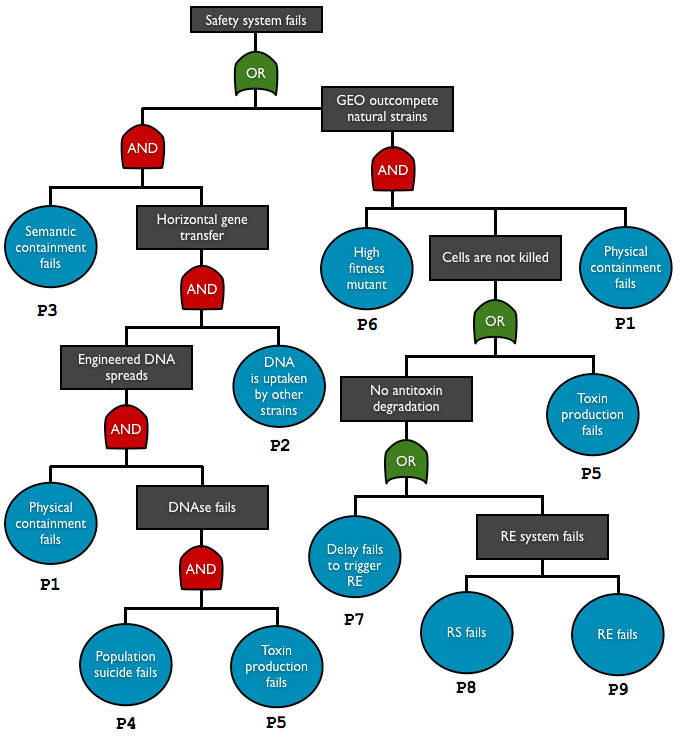
| + | <td style="border:0.035cm solid #000000;padding:0.176cm;"> P1 | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> physical containment | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> leakage | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> DNA/cell escape | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> [http://2012.igem.org/Team:Paris_Bettencourt/Encapsulation#Cell_Containment_Assay experiment], [http://2012.igem.org/Team:Paris_Bettencourt/Modeling#Physical_containment_failure estimation : 4.10<sup>-6</sup>] | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td style="border:0.035cm solid #000000;padding:0.176cm;"> P2 | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> DNA uptake | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> DNA transferred into natural strain | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> HGT | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> transconjugant to donor ratio in HGT is typically < 10<sup>-5</sup> <sup>[[http://2012.igem.org/Team:Paris_Bettencourt/Modeling#References 4]]</sup>, assume this is the rate per generation | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td style="border:0.035cm solid #000000;padding:0.176cm;"> P3 | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> semantic containment | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> genetic failure | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> HGT | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> With 3 amber mutations, probability to recover at least 50% of efficiency : 10<sup>-18</sup> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td style="border:0.035cm solid #000000;padding:0.176cm;"> P4 | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> population suicide | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> no surrounding cells with enough toxin | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> no death | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> not estimated : consider it as 1 meanwhile | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td style="border:0.035cm solid #000000;padding:0.176cm;"> P5 | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> toxin production | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> genetic failure | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> no DNA degradation (w/o antitoxin), no death (with antitoxin) | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> [http://2012.igem.org/Team:Paris_Bettencourt/Modeling#Genetic_failure estimation] : 10<sup>-6</sup> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td style="border:0.035cm solid #000000;padding:0.176cm;"> P6 | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> mutant fitness | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> beneficial mutation | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> out-competition of the mutant | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> not determined, considered as 1. | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td style="border:0.035cm solid #000000;padding:0.176cm;"> P7 | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> delay system | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> genetic failure | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> no antitoxin degradation | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> [http://2012.igem.org/Team:Paris_Bettencourt/Modeling#Genetic_failure estimation] : 10<sup>-6</sup> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td style="border:0.035cm solid #000000;padding:0.176cm;"> P8 | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> restriction enzyme | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> genetic failure | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> no antitoxin degradation | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> [http://2012.igem.org/Team:Paris_Bettencourt/Modeling#Genetic_failure estimation] : 10<sup>-6</sup> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td style="border:0.035cm solid #000000;padding:0.176cm;"> P9 | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> restriction site | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> genetic failure | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> no antitoxin degradation | ||
| + | </td><td style="border:0.035cm solid #000000;padding:0.176cm;"> 10<sup>-9</sup> | ||
| + | </td></tr></table> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | |||
| + | </table> | ||
==Semantic Containment== | ==Semantic Containment== | ||
| Line 53: | Line 189: | ||
| − | ==All biosafety | + | ==All biosafety parts== |
Include previous parts plus other marked as biosafety only. | Include previous parts plus other marked as biosafety only. | ||
| Line 66: | Line 202: | ||
<img src="https://static.igem.org/mediawiki/2012/8/8d/ParisBettencourtPolyethyleneimineAlginate.png" width="800px"> | <img src="https://static.igem.org/mediawiki/2012/8/8d/ParisBettencourtPolyethyleneimineAlginate.png" width="800px"> | ||
</html> | </html> | ||
| + | |||
| + | |||
| + | ==Barcode== | ||
Latest revision as of 21:43, 21 April 2020
Contents
More information about biosafety
- Quick links of pubmed research :
- [http://www.ncbi.nlm.nih.gov/pubmed?term=biosafety%20gmo Biosafety and GMO's]
- [http://www.ncbi.nlm.nih.gov/pubmed?term=biosafety%20synthetic%20biology Biosafety in Synthetic Biology]
- [http://www.ncbi.nlm.nih.gov/pubmed?term=issue%20synthetic%20biology Issues of Synthetic Biology]
We encourage you to do this kind of research as part of your bibliography for whatever project you are working on.
- Here is the link to an openwetware page that should be improved by the community. This report exists to improve biosafety practices in synthetic biology, raise good questions, and encourage collaboration in exploring the issue.
-> [http://openwetware.org/wiki/How_safe_is_safe_enough:_towards_best_pratices_of_synthetic_biology How safe is Safe Enough?]
- Link to the biosafety recommendation page of iGEM:
It includes book, papers and other useful references.
Recommendation from Human Practice Teams
- Report of Sara Aguiton : [http://2009.igem.org/Team:Paris/Ethics_ethicalreport#top report]
- Report of Claire Mayer : [http://2012.igem.org/Team:Paris_Bettencourt/Human_Practice/Report report]
|
Conclusion of these reports :
|
Proposals to improve this page (Feel free to add ideas)
- Add a biosafety grade, that could quantify the degree of robustness. For instance, in semantic containment with amber mutation, we score the robustness with the number of amber mutations.
Example of biosafety systems
Here are reported biosafety systems that teams created. The risk and hazards of these systems are assessed through fault tree analysis, a top-down approach used in engineering field.
| Team | Fault Tree Analysis | Guesstimation | ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Paris Bettencourt 2012 |
|
Semantic Containment
Here are parts that would add a semantic containment to genes, or parts that would allow the cell to read semantic containments gene.
| Name | Type | Description | Length |
|---|---|---|---|
| BBa_K1479017 | Protein_Domain | just for test | 100 |
| BBa_K1479018 | Protein_Domain | Just for test | |
| BBa_K2086001 | Coding | Serine Repeat Antigen (SerA) | 1233 |
| BBa_K914000 | RNA | pLac-supD-T | 241 |
| BBa_K914009 | Translational_Unit | P1003* Ser133->Amber Codon | 967 |
| BBa_K914011 | Intermediate | supD-T - intermediate | 178 |
| BBa_K914012 | Plasmid_Backbone | pSB1A2 with 1 amber mutation | 2079 |
| BBa_K914018 | Translational_Unit | P1003** Kan resistant gene with 2 Amber Codon | 967 |
Kill switch
Here are parts that could trigger cell death through disruption at the level of the protein and the cell.
| Name | Type | Description | Length |
|---|---|---|---|
| BBa_K1074011 | Composite | PsdpRI+RBS+SDP | 2207 |
| BBa_K117000 | Coding | Lysis gene (promotes lysis in colicin-producing bacteria strain) | 144 |
| BBa_K1172901 | Coding | Alanine racemase from ''E. coli'' | 1080 |
| BBa_K1172902 | Device | alanine racemase (''alr'') under the control of the P''tac'' promoter | 1134 |
| BBa_K1172908 | Device | Part 1 of the Biosafety-System Lac of Growth (pRha lacI alr) | 2509 |
| BBa_K1172909 | Device | Biosafety-System araCtive (AraC) | 3281 |
| BBa_K1172911 | Device | Biosafety-System Lac of Growth (lacI) | 3600 |
| BBa_K1172912 | Device | Part 1 of the Biosystem TetOR alive (pRha TetR alr) | 2042 |
| BBa_K1184000 | Coding | KillerRed | 723 |
| BBa_K1197007 | Composite | IPTG dependent B.subtilis Kill Switch construct | 2006 |
| BBa_K1223001 | Project | P.A.S.E. 1 cassette | 2148 |
| BBa_K1223002 | Project | P.A.S.E. 2 cassette | 1210 |
| BBa_K1405008 | Composite | A Kill Switch with "memory" time repressed by IPTG | 2926 |
| BBa_K1616005 | Coding | Holin/endolysin reversed - RBS and double T7 terminator | 1379 |
| BBa_K1616013 | Coding | ccdB reversed - RBS and double T7 terminator | 575 |
| BBa_K1639022 | Composite | pCons-TetR-pTet-TevProtease Suicide Switch | 1546 |
| BBa_K1767001 | Composite | P(Lac)IQ RBS Aiia RBS LuxR term term luxpR | 1818 |
| BBa_K1767002 | Composite | P(Lac)IQ RBS Aiia RBS LuxR ter ter luxpR RBS tetR ter ter | 2666 |
| BBa_K1767003 | Composite | P(Lac)IQ RBS Aiia RBS LuxR ter ter luxpR RBS tetR ter ter P(tetR) RBS mRFP1 ter ter | 3597 |
| BBa_K1767004 | Composite | P(Lac)IQ RBS LuxR ter ter luxpR RBS tetR ter ter | 1892 |
| BBa_K1819008 | Device | Promoter test circuit to analyze the kill switch efficiency | 2725 |
| BBa_K1820024 | Generator | Phage inducible promoter 1997 +RBS+FspI + term | 1160 |
| BBa_K1820025 | Generator | p2001-native RBS-Fsp1-B0015 | 1209 |
| BBa_K1914000 | Coding | E. coli Codon Optimised KillerOrange(EOKO) | 717 |
| BBa_K1914001 | Device | pT7- E. coli optimised - KillerOrange (EOKO) | 926 |
| BBa_K1914003 | Device | pT7- E. coli optimised - KillerRed (EOKR) | 929 |
| BBa_K1919100 | Device | The device consist of temperature promotor and hokD coding gene. | 1112 |
| BBa_K2040122 | Coding | KillerRed + NLS | 744 |
| BBa_K2086002 | Composite | PyeaR - SerA | 1499 |
| BBa_K2150103 | Coding | 1204 toxin CDS | 507 |
| BBa_K2150104 | Coding | 6249 toxin CDS | 319 |
| BBa_K2232025 | Composite | mazEF switch | 1006 |
| BBa_K2292002 | Coding | Toxin-antitoxin mazEF | 584 |
| BBa_K2292006 | Composite | MazEF kill switch | 877 |
| BBa_K2486026 | Composite | pGadA-MazF kill switch | 769 |
| BBa_K2616002 | Coding | Temperature Kill Switch | 938 |
| BBa_K2660006 | Coding | N-Cas9 | 3459 |
| BBa_K2660007 | Coding | C-Cas9 | 648 |
| BBa_K2757000 | Composite | Art-175 fused with N-terminal DsbA 2-19 signal peptide and unidirectional pTet | 1008 |
| BBa_K2757001 | Composite | Art-175 fused with N-terminal DsbA 2-19 signal peptide and bidirectional pTet | 1065 |
| BBa_K2817006 | Composite | PcspA-RBS-mazF | 535 |
| BBa_K2896001 | Coding | Thylakoid-bound SuperNova (for S. elongatus PCC 7942) | 846 |
| BBa_K2919000 | DNA | ghoT toxin from E. coli K12 | 174 |
| BBa_K302034 | Translational_Unit | mazEF toxin-antitoxin cluster | 607 |
| BBa_K302035 | Device | Sucrose-limitation induced kill switch | 1006 |
| BBa_K3036004 | Composite | Cold triggered kill switch | 649 |
| BBa_K3036008 | Coding | RelB with degradation-promoting tag RepA | 1758 |
| BBa_K3096052 | RNA | CRISPR Array targeting the Plasmid DNA of the glucose dependent Exendin-4 expression system | 147 |
| BBa_K3096054 | Composite | Cas3 Nuclease under the control of the AraC promoter pBAD | 2803 |
| BBa_K3096055 | Composite | Type I CRISPR/Cas system surveillance complex Cascade ABCDE under the control of the AraC promoter | 4487 |
| BBa_K3190601 | Composite | pGAL1-BI-I: BI-I CDS under inducible promoter | 1181 |
| BBa_K3215013 | Composite | Tse2 Repressed by Arabinose Kill Switch | 1950 |
| BBa_K3215016 | Composite | MazF repressed by IPTG Kill Switch | 2886 |
| BBa_K3237029 | Composite | nucA | 1754 |
| BBa_K3273001 | Coding | Serratia marcescens nucA nuclease | 801 |
| BBa_K3273008 | Composite | Composite ADH2/TDH3 nucA | 1849 |
| BBa_K3273009 | Composite | Composite HXT6 Bax | 2039 |
| BBa_K3273010 | Composite | Composite MET 25 nucA | 1113 |
| BBa_K3317053 | Coding | NucD | 540 |
| BBa_K3317059 | Composite | NucE lysis construction | 308 |
| BBa_K3317060 | Composite | NucD | 572 |
| BBa_K3317061 | Composite | tetR repressed killswitch | 1504 |
| BBa_K3378002 | Regulatory | Ferrous iron sensetive promoter fhuA1 | 55 |
| BBa_K3512001 | Coding | ccdB Toxin | 306 |
| BBa_K3588014 | Composite | MazE-MazF Hypoxia induced kill switch | 2211 |
| BBa_K3634012 | Composite | Glucose-Mediated Death Sensor | 566 |
| BBa_K3755043 | RNA | Sok RNA | 66 |
| BBa_K3755045 | Coding | Hok/Mok protein | 217 |
| BBa_K3755046 | Composite | Double Regulated(trp+lac) Kill Switch | 2352 |
| BBa_K3848004 | Composite | Lactate regulated kill switch | 2837 |
| BBa_K4133006 | Coding | Repressor protein CtsR_Lp (Lactobacillus plantarum WCFS1) for Lactobacillus casei | 468 |
| BBa_K4133007 | Regulatory | PclpC_Lp repressible promoter (Lactobacillus plantarum WCFS1) | 59 |
| BBa_K4133008 | RBS | RBS_Lp (Lactobacillus plantarum WCFS1) | 14 |
| BBa_K4133009 | Coding | Repressor protein CtsR_Lc (Lactococcus lactis) for Lactobacillus casei | 456 |
| BBa_K4133010 | Regulatory | PclpC_Lc repressible promoter (Lactococcus lactis) | 72 |
| BBa_K4133011 | RBS | RBS_Lc (Lactococcus lactis) | 12 |
| BBa_K4133012 | Coding | LysKB317 (Endolysin) for Lactobacillus casei | 894 |
| BBa_K4202000 | Coding | mazE(codon optimization for Bacillus subtilis) | 249 |
| BBa_K4202001 | Coding | mazF(codon optimization for Bacillus subtilis) | 421 |
| BBa_K4202045 | Generator | Sucrose regulated kill switch in Bacillus subtilis | 1291 |
| BBa_K4345020 | Composite | 5' UTR of hsp17 with ccdA fused to sfGFP | 1197 |
| BBa_K4345023 | Composite | 5' UTR of rpoH with ccdA fused to sfGFP | 1395 |
| BBa_K4345024 | Composite | 5' UTR of prfA with ccdA fused to sfGFP | 1277 |
| BBa_K515104 | Composite | J23100 promoter - Antiholin | 488 |
| BBa_K515106 | Composite | J23103 promoer - RBS B0034-RFP E1010 - Holin K112805 - endolysin K112806 | 1973 |
| BBa_K5480009 | Composite | Y38-DpnI-TrrnB | 2353 |
| BBa_K628006 | Composite | Protegrin-1 Kill Switch | 353 |
| BBa_K879917 | Plasmid_Backbone | LacO ori | 2072 |
| BBa_K879918 | Plasmid_Backbone | TetO ori | 2074 |
XNase
Here are parts that could trigger cell death through disruption at the level of the DNA or RNA.
| Name | Type | Description | Length |
|---|---|---|---|
| BBa_K1172904 | Coding | Rnase Ba (Barnase) from Bacillus amyloliquefaciens | 339 |
| BBa_K131000 | Generator | ColE2 Operon | 2640 |
| BBa_K1479016 | Promoter | -- No description -- | |
| BBa_K1479017 | Protein_Domain | just for test | 100 |
| BBa_K150009 | Generator | ColicinE1 Producer Controlled by 3OC6HSL Receiver Device | 3180 |
| BBa_K526001 | Coding | ptrc*-Dpn | 1552 |
| BBa_K914001 | Composite | pLac Colicin E2 Immunity protein | 345 |
| BBa_K914002 | Intermediate | repressilator RBS Colicin E2 Immunity protein | 282 |
All biosafety parts
Include previous parts plus other marked as biosafety only.
| Name | Type | Description | Length |
|---|---|---|---|
| BBa_K1074011 | Composite | PsdpRI+RBS+SDP | 2207 |
| BBa_K117000 | Coding | Lysis gene (promotes lysis in colicin-producing bacteria strain) | 144 |
| BBa_K1172901 | Coding | Alanine racemase from ''E. coli'' | 1080 |
| BBa_K1172902 | Device | alanine racemase (''alr'') under the control of the P''tac'' promoter | 1134 |
| BBa_K1172904 | Coding | Rnase Ba (Barnase) from Bacillus amyloliquefaciens | 339 |
| BBa_K1172905 | Device | Part 1 of the Biosafety-System araCtive | 2260 |
| BBa_K1172908 | Device | Part 1 of the Biosafety-System Lac of Growth (pRha lacI alr) | 2509 |
| BBa_K1172909 | Device | Biosafety-System araCtive (AraC) | 3281 |
| BBa_K1172911 | Device | Biosafety-System Lac of Growth (lacI) | 3600 |
| BBa_K1172912 | Device | Part 1 of the Biosystem TetOR alive (pRha TetR alr) | 2042 |
| BBa_K1184000 | Coding | KillerRed | 723 |
| BBa_K1197007 | Composite | IPTG dependent B.subtilis Kill Switch construct | 2006 |
| BBa_K1223001 | Project | P.A.S.E. 1 cassette | 2148 |
| BBa_K1223002 | Project | P.A.S.E. 2 cassette | 1210 |
| BBa_K131000 | Generator | ColE2 Operon | 2640 |
| BBa_K1363201 | Composite | Anti-LPS factor(LALF) regulated by IPTG | 2298 |
| BBa_K1405008 | Composite | A Kill Switch with "memory" time repressed by IPTG | 2926 |
| BBa_K1442006 | Regulatory | Anti-theophylline Aptazyme | 113 |
| BBa_K1479005 | Promoter | -- No description -- | 16 |
| BBa_K1479007 | RBS | Just for upload test | 32 |
| BBa_K1479008 | Temporary | short short short | 32 |
| BBa_K1479009 | Temporary | short short short | 32 |
| BBa_K1479011 | Promoter | oh my god | 11 |
| BBa_K1479012 | Temporary | short short short | 32 |
| BBa_K1479013 | Temporary | short short short | 32 |
| BBa_K1479014 | Temporary | Just for Test | 36 |
| BBa_K1479015 | RBS | Just for test | 16 |
| BBa_K1479016 | Promoter | -- No description -- | |
| BBa_K1479017 | Protein_Domain | just for test | 100 |
| BBa_K1479018 | Protein_Domain | Just for test | |
| BBa_K150009 | Generator | ColicinE1 Producer Controlled by 3OC6HSL Receiver Device | 3180 |
| BBa_K1554000 | Regulatory | TA29 promoter | 859 |
| BBa_K1554004 | Device | Yellow biosafety module for plants | 3394 |
| BBa_K1554005 | Device | Blue biosafety module for plants | 3364 |
| BBa_K1616005 | Coding | Holin/endolysin reversed - RBS and double T7 terminator | 1379 |
| BBa_K1616013 | Coding | ccdB reversed - RBS and double T7 terminator | 575 |
| BBa_K1639022 | Composite | pCons-TetR-pTet-TevProtease Suicide Switch | 1546 |
| BBa_K1720002 | Regulatory | Hypoxia-induced promotor | 707 |
| BBa_K1767001 | Composite | P(Lac)IQ RBS Aiia RBS LuxR term term luxpR | 1818 |
| BBa_K1767002 | Composite | P(Lac)IQ RBS Aiia RBS LuxR ter ter luxpR RBS tetR ter ter | 2666 |
| BBa_K1767003 | Composite | P(Lac)IQ RBS Aiia RBS LuxR ter ter luxpR RBS tetR ter ter P(tetR) RBS mRFP1 ter ter | 3597 |
| BBa_K1767004 | Composite | P(Lac)IQ RBS LuxR ter ter luxpR RBS tetR ter ter | 1892 |
| BBa_K1774000 | Project | CRISPR-Cas9 contaiment device repressed by arabinose and tryptophan | 7191 |
| BBa_K1819008 | Device | Promoter test circuit to analyze the kill switch efficiency | 2725 |
| BBa_K1820024 | Generator | Phage inducible promoter 1997 +RBS+FspI + term | 1160 |
| BBa_K1820025 | Generator | p2001-native RBS-Fsp1-B0015 | 1209 |
| BBa_K1914000 | Coding | E. coli Codon Optimised KillerOrange(EOKO) | 717 |
| BBa_K1914001 | Device | pT7- E. coli optimised - KillerOrange (EOKO) | 926 |
| BBa_K1914003 | Device | pT7- E. coli optimised - KillerRed (EOKR) | 929 |
| BBa_K1919100 | Device | The device consist of temperature promotor and hokD coding gene. | 1112 |
| BBa_K2040122 | Coding | KillerRed + NLS | 744 |
| BBa_K2065003 | Coding | CT52-Caspase9 | |
| BBa_K2086001 | Coding | Serine Repeat Antigen (SerA) | 1233 |
| BBa_K2086002 | Composite | PyeaR - SerA | 1499 |
| BBa_K2107001 | Generator | Stationary Phase Promoter + Kil Protein | 263 |
| BBa_K2150102 | Coding | 136 toxin (yobR) CDS | 744 |
| BBa_K2150103 | Coding | 1204 toxin CDS | 507 |
| BBa_K2150104 | Coding | 6249 toxin CDS | 319 |
| BBa_K2150110 | Device | LacI (constitutive) + Ptac + RiboJ + 1204 toxin | 2040 |
| BBa_K2152004 | Other | Bacteriophage Phi X 174 lysis gene E with T7 and RBS | 363 |
| BBa_K2232025 | Composite | mazEF switch | 1006 |
| BBa_K2292002 | Coding | Toxin-antitoxin mazEF | 584 |
| BBa_K2292006 | Composite | MazEF kill switch | 877 |
| BBa_K2365508 | Composite | Bax induced part | 1027 |
| BBa_K2384009 | Regulatory | TetR | 847 |
| BBa_K2384010 | Regulatory | LacI | 1427 |
| BBa_K2384011 | DNA | mf-Lon | 2619 |
| BBa_K2384014 | Composite | Safeguard-toggle switch | 5004 |
| BBa_K2486026 | Composite | pGadA-MazF kill switch | 769 |
| BBa_K2486027 | Coding | MazF endoribonuclease | 339 |
| BBa_K2516003 | Coding | Membrane-bound SuperNova (for C. reinhardtii) | 969 |
| BBa_K2616002 | Coding | Temperature Kill Switch | 938 |
| BBa_K2660006 | Coding | N-Cas9 | 3459 |
| BBa_K2660007 | Coding | C-Cas9 | 648 |
| BBa_K2757000 | Composite | Art-175 fused with N-terminal DsbA 2-19 signal peptide and unidirectional pTet | 1008 |
| BBa_K2757001 | Composite | Art-175 fused with N-terminal DsbA 2-19 signal peptide and bidirectional pTet | 1065 |
| BBa_K2817006 | Composite | PcspA-RBS-mazF | 535 |
| BBa_K2896001 | Coding | Thylakoid-bound SuperNova (for S. elongatus PCC 7942) | 846 |
| BBa_K2919000 | DNA | ghoT toxin from E. coli K12 | 174 |
| BBa_K302034 | Translational_Unit | mazEF toxin-antitoxin cluster | 607 |
| BBa_K302035 | Device | Sucrose-limitation induced kill switch | 1006 |
| BBa_K3036004 | Composite | Cold triggered kill switch | 649 |
| BBa_K3036008 | Coding | RelB with degradation-promoting tag RepA | 1758 |
| BBa_K3096052 | RNA | CRISPR Array targeting the Plasmid DNA of the glucose dependent Exendin-4 expression system | 147 |
| BBa_K3096054 | Composite | Cas3 Nuclease under the control of the AraC promoter pBAD | 2803 |
| BBa_K3096055 | Composite | Type I CRISPR/Cas system surveillance complex Cascade ABCDE under the control of the AraC promoter | 4487 |
| BBa_K3190601 | Composite | pGAL1-BI-I: BI-I CDS under inducible promoter | 1181 |
| BBa_K3215013 | Composite | Tse2 Repressed by Arabinose Kill Switch | 1950 |
| BBa_K3215016 | Composite | MazF repressed by IPTG Kill Switch | 2886 |
| BBa_K3237029 | Composite | nucA | 1754 |
| BBa_K3273001 | Coding | Serratia marcescens nucA nuclease | 801 |
| BBa_K3273008 | Composite | Composite ADH2/TDH3 nucA | 1849 |
| BBa_K3273009 | Composite | Composite HXT6 Bax | 2039 |
| BBa_K3273010 | Composite | Composite MET 25 nucA | 1113 |
| BBa_K3317053 | Coding | NucD | 540 |
| BBa_K3317059 | Composite | NucE lysis construction | 308 |
| BBa_K3317060 | Composite | NucD | 572 |
| BBa_K3317061 | Composite | tetR repressed killswitch | 1504 |
| BBa_K3378002 | Regulatory | Ferrous iron sensetive promoter fhuA1 | 55 |
| BBa_K3512001 | Coding | ccdB Toxin | 306 |
| BBa_K3588014 | Composite | MazE-MazF Hypoxia induced kill switch | 2211 |
| BBa_K3634012 | Composite | Glucose-Mediated Death Sensor | 566 |
| BBa_K3634020 | Coding | CviJI Endonuclease (+ ssrA deg. tag) | 873 |
| BBa_K3755043 | RNA | Sok RNA | 66 |
| BBa_K3755045 | Coding | Hok/Mok protein | 217 |
| BBa_K3755046 | Composite | Double Regulated(trp+lac) Kill Switch | 2352 |
| BBa_K3848004 | Composite | Lactate regulated kill switch | 2837 |
| BBa_K4133006 | Coding | Repressor protein CtsR_Lp (Lactobacillus plantarum WCFS1) for Lactobacillus casei | 468 |
| BBa_K4133007 | Regulatory | PclpC_Lp repressible promoter (Lactobacillus plantarum WCFS1) | 59 |
| BBa_K4133008 | RBS | RBS_Lp (Lactobacillus plantarum WCFS1) | 14 |
| BBa_K4133009 | Coding | Repressor protein CtsR_Lc (Lactococcus lactis) for Lactobacillus casei | 456 |
| BBa_K4133010 | Regulatory | PclpC_Lc repressible promoter (Lactococcus lactis) | 72 |
| BBa_K4133011 | RBS | RBS_Lc (Lactococcus lactis) | 12 |
| BBa_K4133012 | Coding | LysKB317 (Endolysin) for Lactobacillus casei | 894 |
| BBa_K4139000 | Coding | mcyH ABC Transporter for Microcystin Production | 1617 |
| BBa_K4139001 | Composite | mcyH ABC Transporter for Microcystin Production (Composite) | 1796 |
| BBa_K4139002 | Coding | mcyH crRNA for Lbu Cas13a (for E.coli) | 57 |
| BBa_K4139003 | Composite | mcyH crRNA for Lbu Cas13a (Composite) | 219 |
| BBa_K4139004 | Coding | mcyH crRNA for Lbu Cas13a (for Cyanobacteria) | 57 |
| BBa_K4139006 | Coding | mcyH crRNA for Lwa Cas13a (for E.coli) | 75 |
| BBa_K4139007 | Composite | mcyH crRNA for Lwa Cas13a (for E.coli, Composite) | 237 |
| BBa_K4139008 | Coding | mcyH crRNA for Lwa Cas13a (for Cyanobacteria) | 75 |
| BBa_K4202000 | Coding | mazE(codon optimization for Bacillus subtilis) | 249 |
| BBa_K4202001 | Coding | mazF(codon optimization for Bacillus subtilis) | 421 |
| BBa_K4202045 | Generator | Sucrose regulated kill switch in Bacillus subtilis | 1291 |
| BBa_K4345020 | Composite | 5' UTR of hsp17 with ccdA fused to sfGFP | 1197 |
| BBa_K4345023 | Composite | 5' UTR of rpoH with ccdA fused to sfGFP | 1395 |
| BBa_K4345024 | Composite | 5' UTR of prfA with ccdA fused to sfGFP | 1277 |
| BBa_K4706007 | Coding | AtBAG6 induced S. cerevisae cell death | 405 |
| BBa_K4706013 | Composite | Gal1 induced RFP A2 AtBAG6 | 2955 |
| BBa_K4706015 | Composite | SrpR repressable PHO5 phosphate dependent cell death | 3532 |
| BBa_K515104 | Composite | J23100 promoter - Antiholin | 488 |
| BBa_K515106 | Composite | J23103 promoer - RBS B0034-RFP E1010 - Holin K112805 - endolysin K112806 | 1973 |
| BBa_K526001 | Coding | ptrc*-Dpn | 1552 |
| BBa_K5480009 | Composite | Y38-DpnI-TrrnB | 2353 |
| BBa_K628006 | Composite | Protegrin-1 Kill Switch | 353 |
| BBa_K748007 | Composite | Biofilm formation device. yddV gene with the IPTG inducible promoter. | 1604 |
| BBa_K772101 | Composite | Self-Destruction Kit: Endolysin + Holin | 839 |
| BBa_K786031 | Reporter | T7promoter-RSR-T7teminator | 956 |
| BBa_K843004 | Device | T4 lysis device regulated by IPTG | 2871 |
| BBa_K879917 | Plasmid_Backbone | LacO ori | 2072 |
| BBa_K879918 | Plasmid_Backbone | TetO ori | 2074 |
| BBa_K914000 | RNA | pLac-supD-T | 241 |
| BBa_K914001 | Composite | pLac Colicin E2 Immunity protein | 345 |
| BBa_K914002 | Intermediate | repressilator RBS Colicin E2 Immunity protein | 282 |
| BBa_K914009 | Translational_Unit | P1003* Ser133->Amber Codon | 967 |
| BBa_K914011 | Intermediate | supD-T - intermediate | 178 |
| BBa_K914012 | Plasmid_Backbone | pSB1A2 with 1 amber mutation | 2079 |
| BBa_K914018 | Translational_Unit | P1003** Kan resistant gene with 2 Amber Codon | 967 |
| BBa_K990005 | Composite | LasR Device + T4 Lysis Device | 2915 |
Physical Containment