Part:BBa_K2152004
Bacteriophage Phi X 174 lysis gene E with T7 and RBS
Lysis protein E spanning the inner and outer membrane of the bacterial, leading to low local degradations of peptidoglycan, allow the release of cytoplasmic content. Use IPTG to induce the transcription of T7 promoter,and the expression of gene E protein.
Usage and Biology
Characterizations of BBa_K2152004 by ShanghaitechChina_2019 team
We made the OD600 of BL21 reach approximately 0.5. Then we added IPTG at the following concentrations: 0 mM, 0.1 mM, 0.5 mM and 1.0 mM. Besides, to ensure that the suicide effect is not caused by the toxicity of IPTG, we added groups of BL21 which contain only pET28(a) backbone without GeneE as the control. We monitored the OD600 of BL21 every 1 hour for 6 hours after addition of IPTG and the final results are shown in the graph below. In the graph, the blue lines show the OD600 of the control groups and the pink ones show the experiment groups. The deeper the color, the higher the concentration of IPTG. The lightest pink is an experiment group with no IPTG. We can see that although higher IPTG concentrations reduce the growth rate when compared to IPTG = 0 group, geneE expression has an obvious killing effect on the experiment groups as the OD600 decreases in 6 hours.
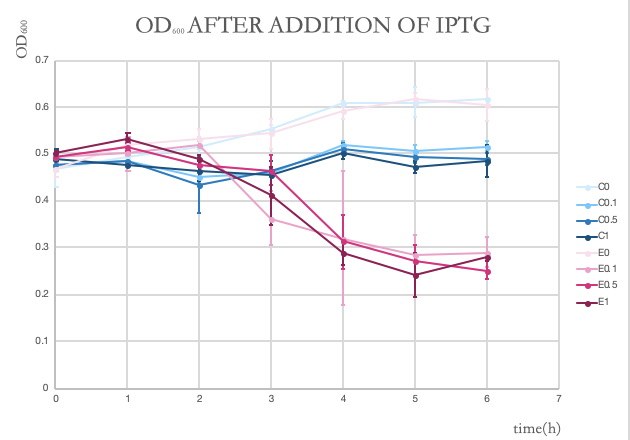
This graph shows the OD600 changes after addition of IPTG in six hours. Pink lines stand for experiment groups (with GeneE) and blue ones stand for control groups (without GeneE). Different numbers stand for different concentrations of IPTG: C0 - control group with no IPTG; C0.1 - control group with IPTG of 0.1mM; C0.5 - control group with IPTG of 0.5mM; C1 - control group with IPTG of 1.0mM; E0 - experiment group with no IPTG; E0.1 - experiment group with IPTG of 0.1mM; E0.5 - experiment group with IPTG of 0.5mM; E1 - experiment group with IPTG of 1.0mM.
We also checked the escape frequency of this kill switch. Utilizing plates with and without IPTG respectively, we can count the bacterial colony number and calculate the escape frequency. We can see in the pictures that the 10-7 dilution drop in the IPTG(+) groups have fewer colonies that IPTG(-) groups. We calculated the escape frequency by this formula:
EscapeFrequency= (Coloniesonnonpermissiveplate x dilution)/ (Coloniesonpermissiveplate x dilution). The result calculated is 12.02%.
IPTG(+) groups are plates with IPTG, IPTG(-) groups are plates without IPTG. In the picture above, we can clearly see the difference in the red circled 10-7 groups where IPTG(+) group only has 1 bacterial colony while IPTG(-) group has over 10 colonies.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
we construct the gene circuit, putting gene E downstream T7 promoter, and induce the expression of gene E under small chemistry molecular, IPTG. These diagram below illustrate the efficiency of gene E which is under T7 promoter. Under the induction of the IPTG, the gene E can strongly inhibit the growth of E.coli.
Figure 8. The growth curve of gene E under T7 promoter which was induced in the time 0, showing the effecient of gene E
Figure 9. The difference of growth curve of gene E under T7 promoter which indicate the efficient of gene E
On the other hand, we found that, even when we use the BL21(DEe)pLysS, which could express toxin protein, the results showed some leaking expression, which make it’s growth curve below that of control group (which contains the same vector without gene E). However, the effect of gene E in experiment group can be seen clearly after the addition of IPTG.
| None |

