Part:BBa_K3215016
MazF repressed by IPTG Kill Switch
This part counts as an improvement of the "A Kill Switch with "memory" time repressed by IPTG" part (BBa_K1405008). Since that for a kill switch we don't want glucose interference in the cell response to IPTG, we removed the CRP DNA binding site of the promoter and inserted a non-responding one, so that the organism won't be influenced by the presence of glucose. Nonetheless, we also removed an operator region controlling cI transcription, in order to loosen cI regulation.
An improved part
Background
During the development of our project, we always considered biosafety as one of our major concerns. Designing a bacteria able to degrade a compound of interest is not enough; we need to make sure it won’t damage an already well established natural environment, which could be a consequence of bacterial leakage.
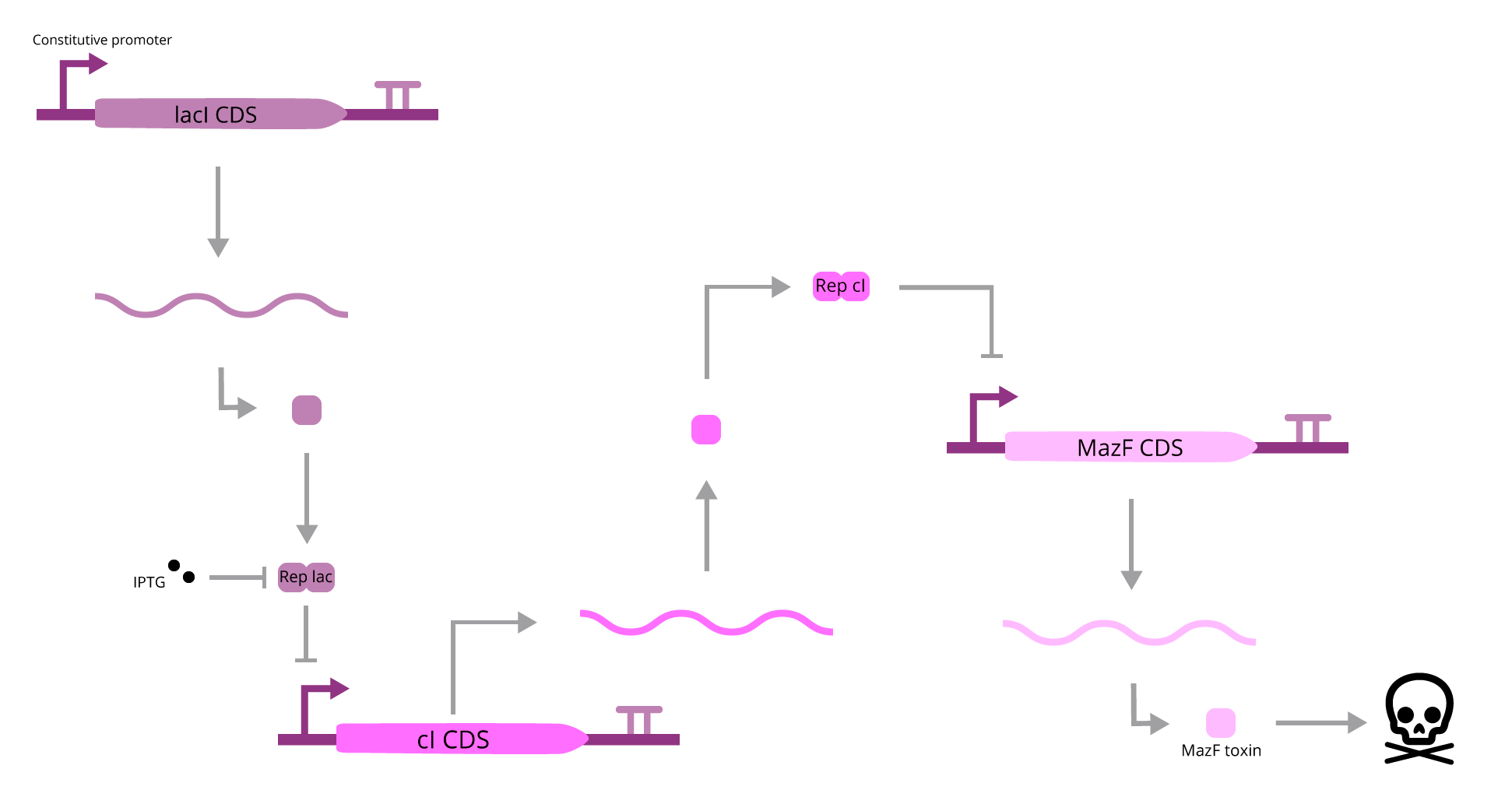
The part designed by Team 2014 BNU China (BBa_K1405008), A Kill Switch with “memory” time repressed by IPTG, assumes that when IPTG is present, it will bind to lac Repressor, which will promote cI transcription. When cI is present, it will bind to the operator region downstream of its CDS, inhibiting the expression of MazF. If IPTG is absent, this mechanism won’t work, and MazF production will be activated, leading to cell death. Although coherent, this explanation lacks one major component: cAMP. When cAMP is present (in a low glucose media), it will activate cI transcription, interfering with IPTG mediated regulation.
Design
To avoid this interference, we must remove the cAMP dependence on the system, and to help IPTG function, we can loosen cI expression regulation, since if it’s much tight, it will be hard for its levels to rise, leading to a leaky transcription of MazF, event that could kill the cell.
To do so, our team faced two other problems, which are that all of the CDSs of this system lack a RBS (sequences will not be translated), and that the cI CDS has a barcode sequence, which Team 2010 UC Davis showed to be problematic, since it activates downstream transcription. To correct these issues, a new BioBrick was created (BBa_K3215014), but we do not consider it as an improvement, since we assumed that those flaws to be extrinsic to the parts idea.
At this point, our team tried to solve two issues: remove cAMP regulation and loosen cI transcription. In order to achieve that, a mathematical model was devised, which all details can be found within section 2.1 of our modelling page. To compare the efficiency of the BioBrick to be improved, we created this BioBrick (BBa_K3215016), which lacks CRP DNA binding site and lacks one of the lac Repressor operators (therefore depleting cAMP/glucose stimulation and facilitating cI transcription). The comparison was made with another model, which can also be found in our modelling page (section 2.2).
Results: MazF
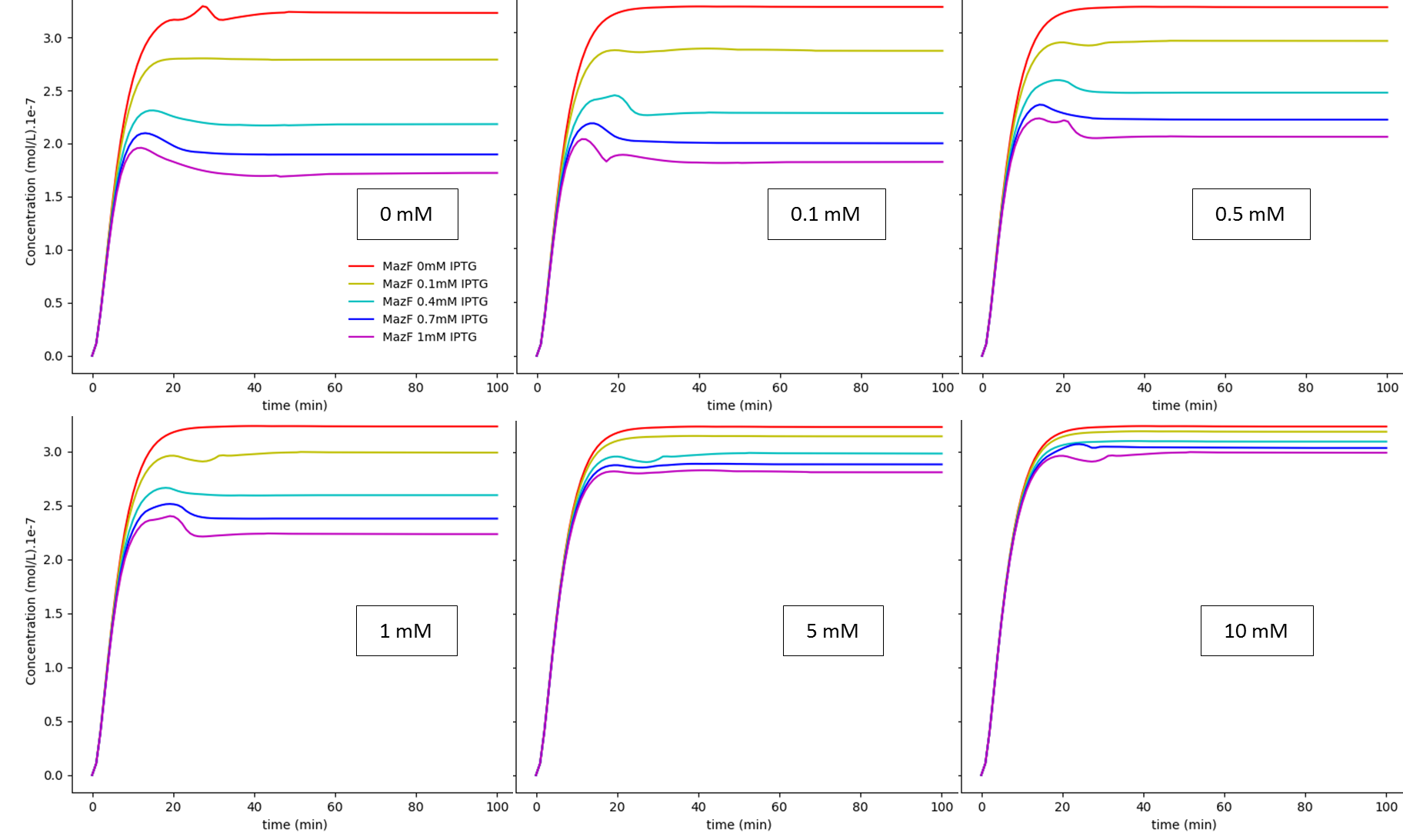
Fig. 1. "A Kill Switch with "memory" time repressed by IPTG": IPTG regulation dependence on glucose concentration: MazF. The values shown inside each box are the extracellular glucose concentrations.
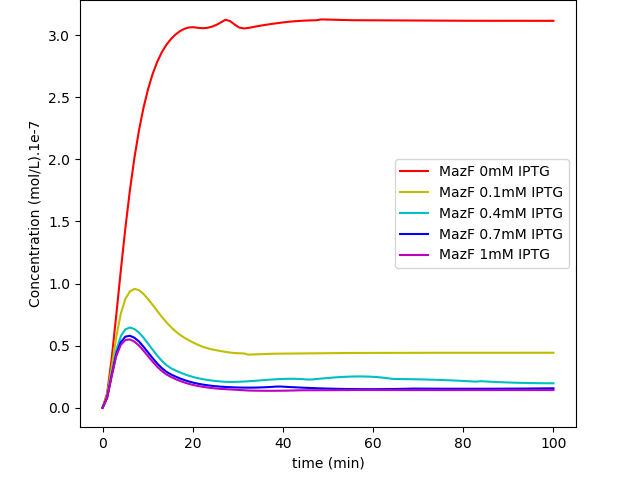
Fig. 2. "MazF repressed by IPTG Kill Switch": MazF levels in a system without glucose regulation and with a loosen cI transcription.
Results: cI
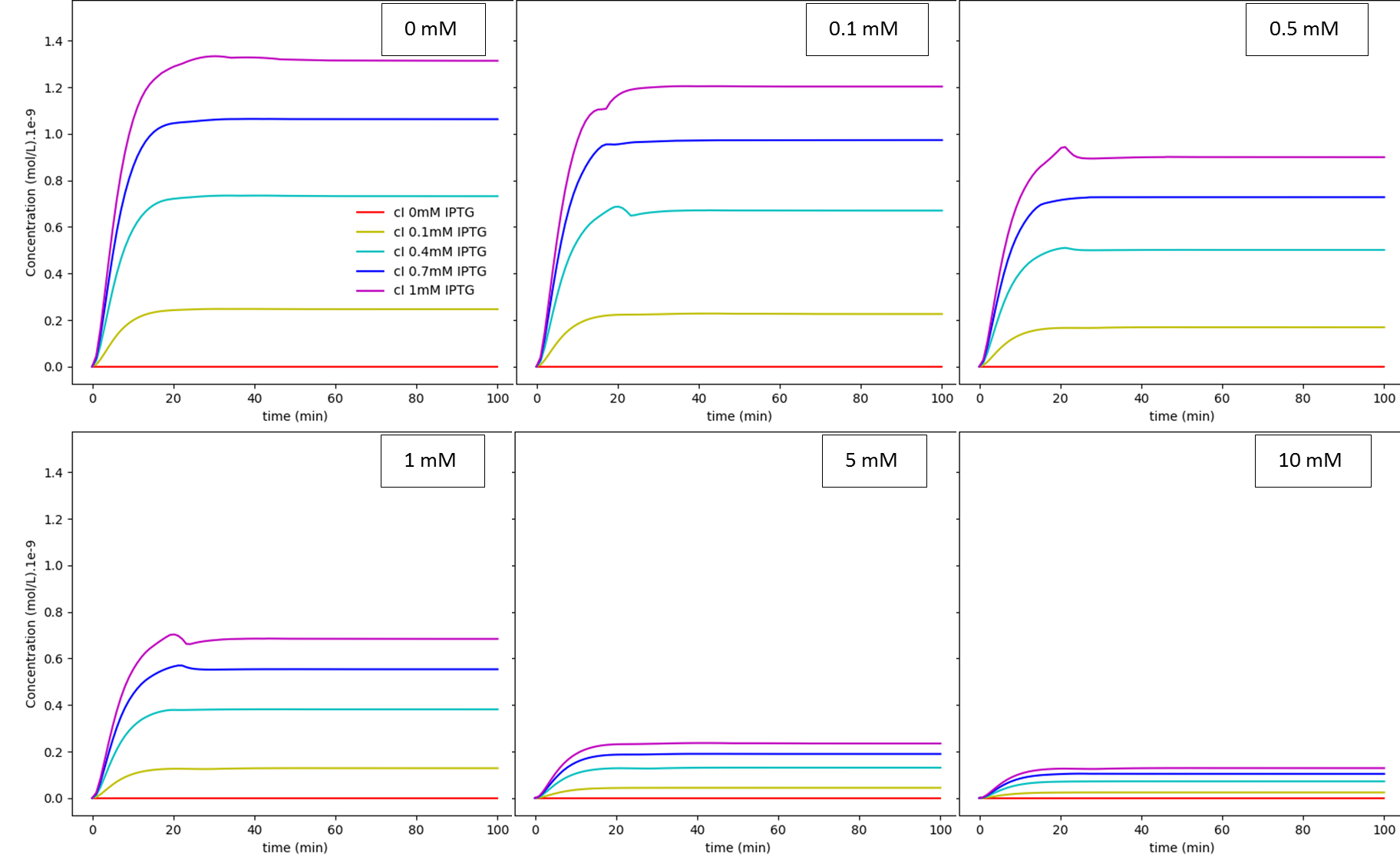
Fig. 3. "A Kill Switch with "memory" time repressed by IPTG": IPTG regulation dependence on glucose concentration: cI. The values shown inside each box are the extracellular glucose concentrations.
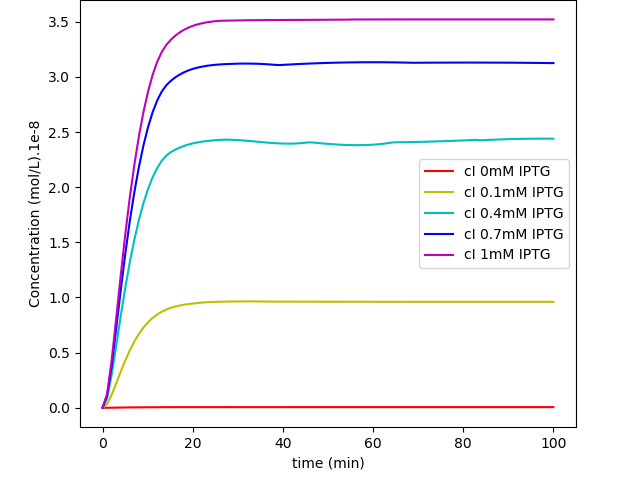
Fig. 4. "MazF repressed by IPTG Kill Switch": cI levels in a system without glucose regulation and with a loosen cI transcription.
Discussion and conclusions
The first system shows that when the concentration of glucose increases, the influence of IPTG on the system decreases, as expected. On the other hand, together, these results show us that besides the absence of glucose interference, this new system has a much higher efficiency when compared to the first system in a natural environment that lacks glucose. Like discussed in our modelling page, the removal of O3 region showed a less rigid cI regulation, and this fact can be in part attributed to a decrease of the “leaky transcriptional inhibition” activity promoted by CRP in the first model - when glucose was absent -, which leads to an attenuation of the levels of cI, and therefore, enhancement of MazF’s.
The major advantage of this new BioBrick is that we can now grow our bacteria in a medium containing glucose, which is way cheaper and easier to produce than other media. We do not consider this “less MazF levels” as an immediate upgrade, for the fact that we don’t know the MazF concentration toxicity threshold established for this system; if this critical value is achieved by inhibition - in low concentrations of glucose - of the last system, one could still use the latter, considering that in a natural environment, the odds to have high levels of glucose (~5mM), which could interfere in bacterial survival, are very low.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1208
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |