Part:BBa_K3273001
Serratia marcescens nucA nuclease
This part is a CDS that produces Serratia marcescens NucA nuclease. This enzyme is capable of cleaving both DNA and RNA, either single or double-stranded. It was implemented in our project as part of a kill switch in order to degrade the transgenic DNA and prevent it from being available for bacteria and other organisms in the environment.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
nucA S. marcescens
Initially planned to be inserted in yeast alongside many other components that are part of our kill switch, we decided to attempt an individual characterization of this part in E. coli. Although we could not express this part in bacterial cells, some milestones were achieved.
CDS Joining
This parts CDS was divided between two gene fragments provided by Twist Bioscience. So the first step was to join it. We achieved this by using fusion PCR and agarose gel DNA purification. The fragments that its CDS was inserted were both part of our overall kill switch, having 1290 and 916 base pairs, with a homology of 92 base pairs between them. This means the fused amplicon should be 2114 base pairs long. A band this size was visible after carrying out the fusion PCR, as shown below in the highlighted sector.
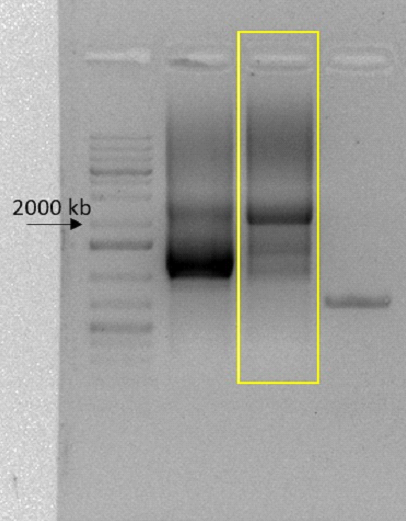
CDS isolation and amplification
With both fragments successfully joined, the next step was to remove the desired CDS from the whole fragment. In order to do it, we designed a pair of primers with tails consisting of restriction enzymes’ restriction site. The forward primer had a BamHI site and the reverse primer had a XhoI restriction site. The primers worked flawlessly, removing the 801 nucleotide CDS from the fused fragment (demonstrated in the image below).

Taq Incubation
In order to try a ligation in pGEM T Easy vector, the amplicon was incubated at 72°C for one hour to add a hangA with Taq polymerase.
Ligation attempt in pGEM T Easy
During the next few days, we tried to ligate the CDS in pGEM T Easy vector, but we were unsuccessful.
Digestion with BamHI and XhoI
Simultaneously with our ligation attempt in pGEM T Easy, we directly digested the amplicon and the expression vector pET-28a with BamHI and XhoI. We had confirmation that the plasmid was linearized, but couldn’t confirm whether both enzymes cut it or only one. Furthermore, we could not verify if the double digestion worked for the amplicon. The image below shows the digested pET-28a and the supposedly digested amplicon (from left to right: marker, undigested pET-28a, digested pET-28a and digested amplicon).
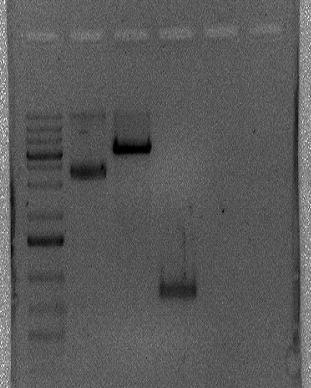
Ligation attempt
During the next few days, we tried a variety of ligation protocols for both pGEM T Easy and pET-28a plasmids, but unfortunately we didn’t obtain positive colonies.
| None |
