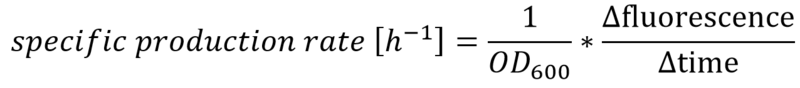Part:BBa_K1172911
Biosafety-System Lac of Growth (lacI)
Usage and Biology
The Biosafety-System Lac of Growth BBa_K1172911 is an improvement of the BioBrick BBa_K914014 by replacing:
- the first promoter PBAD into the rhamnose promoter PRha to obtain a lower basal transcription.
- integration of the alanine racemase (alr) BBa_K1172901 to obtain a higher plasmid stability and taking advantage of the double-kill switch.
For more details about the seperate genes in this Biosafety-System and their function, click [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_L here]
For more details about the seperate genes in this Biosafety-System and their function, click [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_L here]
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1623
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1547
Illegal BamHI site found at 2249 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1665
Illegal AgeI site found at 1965 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1422
Illegal BsaI.rc site found at 3387
Biosafety-System Lac of Growth
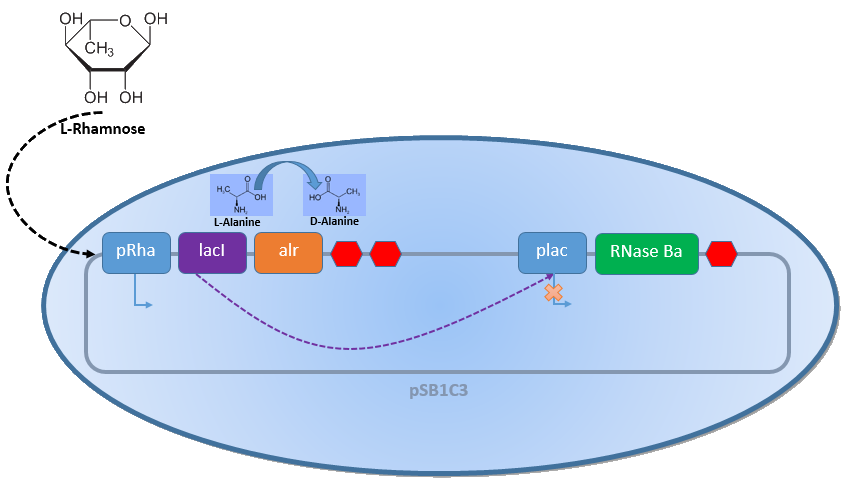
Combining the [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_L genes] described above with the Biosafety-Strain K-12 ∆alr ∆dadX results in a powerful device, allowing us to control the bacterial cell division. The control of the bacterial growth is possible either active or passive. Active by inducing the PLac promoter with IPTG or classically with lactose and passive by the induction of L-rhamnose. The passive control makes it possible to control the bacterial cell division in a defined closed environment, like the MFC, by continuously adding L-rhamnose to the medium. As shown in Figure 1 below, this leads to an expression of the essential alanine racemase (alr) and the LacI repressor, so that the expression of the RNase Ba is repressed.
In the event that bacteria exit the defined environment of the MFC or L-rhamnose is not added to the medium any more, both the expression of the alanine Racemase (Alr) and the LacI repressor decrease, so that the expression of the toxic RNase Ba (Barnase) begins. The cleavage of the intracellular RNA by the Barnase and the lack of synthesized D-alanine, caused by the repressed alanine racemase inhibit the cell division. Through this it can be secured that the bacteria can only grow in the defined area or the device of choice respectively.

Characterization of the lactose promoter Plac
First the lactose promoter PLac was characterized to get a first impression of its basal transcription rate. Therefore the bacterial growth was investigated under the pressure of the unrepressed Plac promoter on different carbon source using M9 minimal medium with either glucose or glycerol. The transcription rate was identified by fluorescence measurement of GFP BBa_E0040 behind the Plac promoter using the BioBrick BBa_K741002.
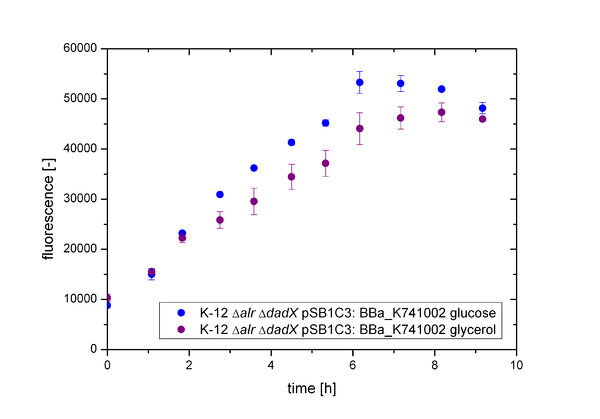
As shown in Figure 3 below, the bacteria grew better on glucose then on glycerol. This is due to glucose being the better energy source of these two, because glycerol enters glycolysis at a later step and therefore delivers less energy. Moreover an additional ATP consumption is needed to drive glycerol uptake. For the investigation of the basal transcription the fluorescence measurements, shown in Figure 4, is more interesting.
It can be demonstrated that the fluorescence differs corresponding to the carbon source used. The difference it not that high as observed for the arabinose promoter [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_S#Characterization_of_the_arabinose_promoter_PBAD PBAD]. But in comparison to the [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_S#Characterization_of_the_arabinose_promoter_TetO tetO] operator the difference is still obvious. This shows that the transcription of the lac promoter depends on the intracellular cAMP level, but is not as effective as the arabinose promoter PBAD. So growth on glucose does not cause an additional induction of the lactose promoter Plac, while growth on glycerol does. In the presence of glucose, the intracellular concentration of cAMP is low. The absence of cAMP results in inactive CAP (carbon utilization activator protein) and thus no induction of more inefficient catabolic routes, like the catabolism of lactose. Therefore, glucose is catabolized first by the bacteria, resulting in an optimal growth. In the absence of glucose, the cAMP level increases which enhances via CAP-cAMP the transcription of most operons encoding enzymes for the catabolism of alternative carbon sources. Therefore, the expression of GFP under the control of the Plac promoter increases on glycerol.

The effect that glucose represses the transcription of the Plac promoter becomes more obvious by calculating the specific production rates of GFP, as shown in Figure 5. The specific production rates were calculated via equation (1) :
With the calculation of the specific production rate of GFP it can be demonstrated that the GFP synthesis rates differ between the cultivation on glucose and glycerol. The specific production rate is low, when using glucose as carbon source, but shows a higher more unstable level in the cultivation with glycerol.
Because the specific production rate was calculated between every single measurement point, the curve in Figure 11 is not smoothed and so the fluctuations have to be ignored, as they do not stand for real fluctuations in the expression of GFP. They are caused by measurement variations in the growth curve and the fluorescence curve. And as neither of this curves are ideal, the fluctuations are the result. Nevertheless this graph shows the difference between the two carbon sources.
Characterization of the Biosafety-System Lac of Growth
The Biosafety-System Lac of Growth was characterized on M9 minimal medium using glycerol as carbon source. As for the characterization of the pure lactose promoter Plac above, the bacterial growth and the fluorescence of GFP BBa_E0040 was measured. Therefore, the wild type and the Biosafety-Strain E. coli K-12 ∆alr ∆dadX both containing the Biosafety-Plasmid BBa_K1172911 were cultivated once with the induction of 1% L-rhamnose and once only on glycerol.
It becomes obvious (Figure 6) that the bacteria, induced with 1 % L-rhamnose (red and black curve) grow obviously slower, than on pure glycerol (orange and blue curve). This might be attributed to the high metabolic burden encountered by the induced bacteria. The expression of the repressor LacI and the alanine racemase (Alr) simultaneously causes a high stress on the cells, so that they grow slower than the uninduced cells which express only GFP. Additionally, it can be observed, that the Biosafety-Strain E. coli K-12 ∆alr ∆dadX shows a very long lag-phase.
The long lag-phase and the high measured fluorescence can not be explained so far, as the wild type shows normal grow. So further cultivation of the Biosafety-Strain containing the Biosafety-System Lac of Growth are necessary. In contrast the fluorescence of the wild type containing the Biosafety-System Lac of Growth shows the expected characteristics. It can be observed that the fluorescence of GFP increases in the cultivation on pure glycerol, while it is reduced by the induction of 1% L-rhamnose.


From the data presented above it cannot be determined if the expression of the repressor LacI does affect the transcription of GFP or not. Considering the wild type containing the Biosafety-System, the slower growth is a first indication that the repressor lacI and the alanine racemase (Alr) are highly expressed, but the growth of the bacteria shows nearly the same kinetics as the fluorescence. So it could be possible that the repressor does not affect the expression level of GFP under the control of the lactose promoter Plac. This becomes more clear by the calculation of the specific production rate of GFP by equation (1) . As shown in Figure 8 below the specific production rate differs between the uninduced Biosafety-System and the Biosafety-System induced by 1% L-rhamnose.
The production of GFP in the presence of L-rhamnose (red curve) is always lower than in its absence (orange curve), so that the expression of GFP is repressed in the presence of L-rhamnose.
Only at the end, the GFP synthesis of the uninduced cultivations seems to be lower. This might be caused by the very fast cell division in the end of the exponential phase, so that cells grow much faster, than expressing GFP.
Because the specific production rate of GFP was calculated between every single measurement point, the curve in Figure 14 is not smoothed and so the fluctuations have to be ignored, as they do not stand for real fluctuations in the expression of GFP. They are caused by measurement variations in the growth curve and the fluorescence curve. But there is a tendency that the production of GFP is lower when the bacteria are induced with 1% L-rhamnose. So the Biosafety-System Lac of Growth seems to work.
Conclusion of the Results
[http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_L#Conclusions see here]
References
- Baldoma L and Aguilar J (1988) Metabolism of L-Fucose and L-Rhamnose in Escherichia coli: Aerobic-Anaerobic Regulation of L-Lactaldehyde Dissimilation [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC210658/pdf/jbacter00179-0434.pdf|Journal of Bacteriology 170: 416 - 421.].
- Busby S, Ebright RH (1999) Transcription activation by catabolite activator protein (CAP).[http://ac.els-cdn.com/S0022283699931613/1-s2.0-S0022283699931613-main.pdf?_tid=a8f22b74-2cf4-11e3-9457-00000aab0f6c&acdnat=1380891687_f17fd24e5a3e15da96493226fdcaaa10|Journal of molecular biology 239: 199 - 213.].
- Carafa, Yves d'Aubenton Brody, Edward and Claude (1990) Thermest Prediction of Rho-independent Escherichia coli Transcription Terminators - A Statistical Analysis of their RNA Stem-Loop Structures [http://ac.els-cdn.com/S0022283699800059/1-s2.0-S0022283699800059-main.pdf?_tid=ede07e2a-2a92-11e3-b889-00000aab0f6c&acdnat=1380629809_2d1a59e395fc69c8608ab8b5aea842f7|Journal of molecular biology 3: 835 - 858].
- Görke, Boris and Stülke, Jörg (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients [http://www.nature.com/nrmicro/journal/v6/n8/full/nrmicro1932.html|Nature Reviews Microbiology 6: 613 - 624].
- Jacob, F Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. [http://www.ncbi.nlm.nih.gov/pubmed/13718526|Journal of molecular biology 216: 318 - 356].
- Lewis, Mitchell (2005) The lac repressor [http://www.sciencedirect.com/science/article/pii/S1631069105000685|Comptes rendues biologies 328: 521 – 548].
- Mossakowska, Danuta E. Nyberg, Kerstin and Fersht, Alan R. (1989) Kinetic Characterization of the Recombinant Ribonuclease from Bacillus amyloliquefaciens (Barnase) and Investigation of Key Residues in Catalysis by Site-Directed Mutagenesis [http://pubs.acs.org/doi/pdf/10.1021/bi00435a033|Biochemistry 28: 3843 - 3850.].
- Paddon, C. J. Vasantha, N. and Hartley, R. W. (1989) Translation and Processing of Bacillus amyloliquefaciens Extracellular Rnase [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC209718/pdf/jbacter00168-0575.pdf|Journal of Bacteriology 171: 1185 - 1187.].
- Voss, Carsten Lindau, Dennis and Flaschel, Erwin (2006) Production of Recombinant RNase Ba and Its Application in Downstream Processing of Plasmid DNA for Pharmaceutical Use [http://onlinelibrary.wiley.com/doi/10.1021/bp050417e/pdf|Biotechnology Progress 22: 737 - 744.].
- Walsh, Christopher (1989) Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. [http://www.jbc.org/content/264/5/2393.long|Journal of biological chemistry 264: 2393 - 2396.]
- Wickstrum, J.R., Santangelo, T.J., and Egan, S.M. (2005) Cyclic AMP receptor protein and RhaR synergistically activate transcription from the L-rhamnose-responsive rhaSR promoter in Escherichia coli. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1251584/?report=reader|Journal of Bacteriology 187: 6708 – 6719.].
Functional Parameters
| None |