Part:BBa_K2660006
N-Cas9
Coding sequence for the N-terminal section of split Cas9 nuclease.
This BioBrick was engineered to be the N-terminal domain of cas9 (BBa_K2457001) coding sequence from Streptococcus pyogenes. It was designed to be an inactive domain linked with proteins like pMag (BBa_K2660009) to control to control the kinetics of activation by blue light and induce targeted genome sequence modifications (to kill or remove the recombinant DNA). This activity can be switched off simply by extinguishing the light.
The nuclease activity requires the use together with nMag(BBa_K2660008) linked with the c-cas9 domain (BBa_K2660007)
N-Cas9
USTC
This part is inserted into plasmid, and the correct construction of this recombinant plasmid was confirmed by PCR identification and sequencing of the PCR products.
(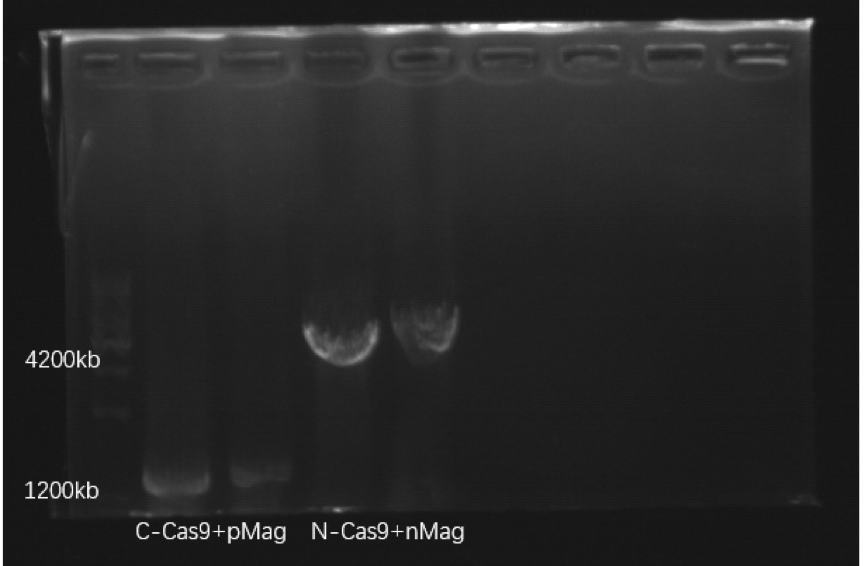 )
)
Figure1. Electrophoresis result PCR of C-Cas9+pMag and N-Cas9+nMag.
Besides, the E.coli was grown in LB liquid medium, and obtain protein by heating them. The sample was electrophoresed on a sodium dodecyl sulfate(SDS)-polyacrylamide gel, followed by Coomassie blue staining.( The pictures are blurred because of the poor photographic equipment)
(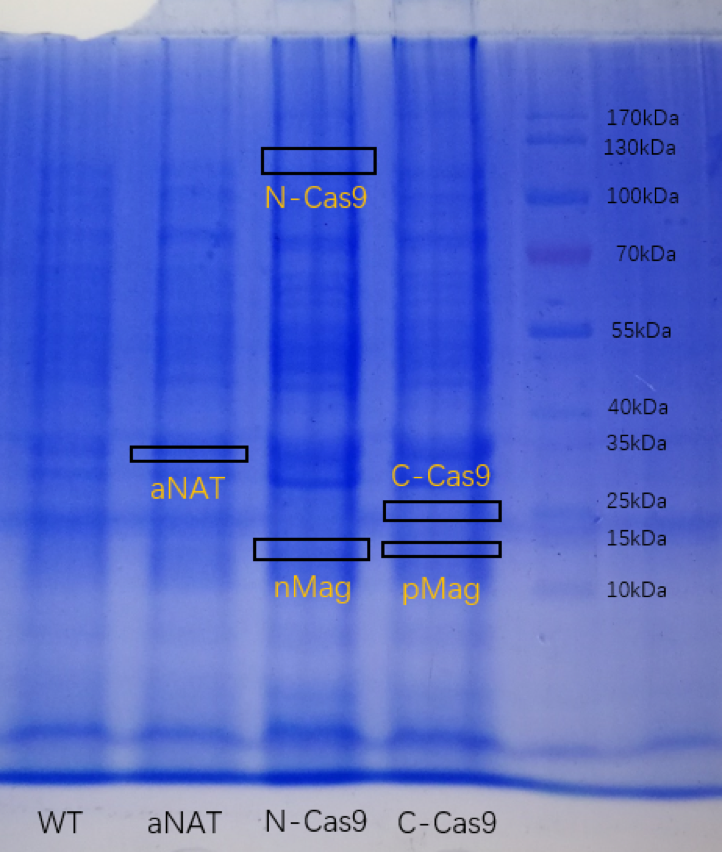 )
)
Figure2. SDS-PAGE for strain expressing aNAT, C-Cas9+pMaga nd N-Cas9+nMag.
references
Nihongaki, Y et al. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nature Biotechnology volume 33, pages 755–760 doi: 10.1038/nbt.3245 (2015)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//chassis/prokaryote/bsubtilis
//collections/probiotics/biocontainment
//function/crispr/cas9
| None |
