Difference between revisions of "Bacillus subtilis"
m |
|||
| (31 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| Ôłĺ | [[ | + | {{Catalog/MainLinks}} |
| + | [[Catalog|< Back to Catalog]] | ||
| Ôłĺ | {{: | + | {{:{{PAGENAME}}/Overview}} |
<html> | <html> | ||
| Line 11: | Line 12: | ||
</html> | </html> | ||
| Ôłĺ | ==Promoters== | + | {| |
| + | |valign='top' align='center' width=50px | {{Click || image=Part icon plasmid backbone.png | link={{PAGENAME}}#Plasmid backbones |width=30px | height=30px}} | ||
| + | |valign='top' |'''[[{{PAGENAME}}#Plasmid backbones|Plasmid backbones]] [[Help:Plasmid backbones|(?)]]''' | ||
| + | |valign='top' align='center' width=50px | {{Click || image=Part icon regulatory.png | link={{PAGENAME}}#Constitutive promoters |width=30px | height=30px}} | ||
| + | |valign='top'|'''[[{{PAGENAME}}#Constitutive promoters|Promoters]] [[Help:Promoters|(?)]]''' | ||
| + | |valign='top' align='center' width=50px | {{Click || image=Part icon rbs.png | link={{PAGENAME}}#Ribosome binding sites |width=30px | height=30px}} | ||
| + | |valign='top' |'''[[{{PAGENAME}}#Ribosome binding sites|Ribosome binding sites]] [[Help:Ribosome Binding Sites|(?)]]''' | ||
| + | |- | ||
| + | |valign='top' align='center' width=50px | {{Click || image=Part icon cds.png | link={{PAGENAME}}#Protein coding sequences |width=30px | height=30px}} | ||
| + | |valign='top' |'''[[{{PAGENAME}}#Protein coding sequences|Protein coding sequences]] [[Help:Protein coding sequences|(?)]]''' | ||
| + | |valign='top' align='center' width=50px | {{Click || image=Part icon dna.png | link={{PAGENAME}}#DNA parts |width=30px | height=30px}} | ||
| + | |valign='top'|'''[[{{PAGENAME}}#DNA parts|DNA parts]] [[Help:DNA|(?)]]''' | ||
| + | |} | ||
| + | |||
| + | {{:{{PAGENAME}}/Help}} | ||
| + | |||
| + | ===Previous iGEM ''Bacillus subtilis'' Projects=== | ||
| + | <html> | ||
| + | <table class="collectionTable sortable"> | ||
| + | <tr> | ||
| + | <th style="width: 120px;">Team</th> | ||
| + | <th style="width: 60px;">Year</th> | ||
| + | <th style="width: 60px;">Parts</th> | ||
| + | <th>Project Title</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2015.igem.org/Team:DTU-Denmark">DTU-Denmark</a></td> | ||
| + | <td>2015</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2015&group=DTU-Denmark">Parts</a></td> | ||
| + | <td class="project">The Synthesizer: Development of antibiotic libraries through Multiplex Automated Genome Engineering</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2015.igem.org/Team:Groningen">Groningen</a></td> | ||
| + | <td>2015</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2015&group=Groningen">Parts</a></td> | ||
| + | <td class="project">Blue Bio Energy</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2015.igem.org/Team:Nagahama">Nagahama</a></td> | ||
| + | <td>2015</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2015&group=Nagahama">Parts</a></td> | ||
| + | <td class="project">''ÚŽÖŔöÁň║ź''ŃÇÇFlavorator: New food preservation method by rose odor E. coli</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2015.igem.org/Team:Nanjing-China">Nanjing-China</a></td> | ||
| + | <td>2015</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2015&group=Nanjing-China">Parts</a></td> | ||
| + | <td class="project">Metallosniper: ┬Śinnovative total solution for heavy metals</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2015.igem.org/Team:NEFU_China">NEFU_China</a></td> | ||
| + | <td>2015</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2015&group=NEFU_China">Parts</a></td> | ||
| + | <td class="project">Yogurt Guarder</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2015.igem.org/Team:Stanford-Brown">Stanford-Brown</a></td> | ||
| + | <td>2015</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2015&group=Stanford-Brown">Parts</a></td> | ||
| + | <td class="project">biOrigami: A New Approach to Reduce the Cost of Space Missions</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2015.igem.org/Team:Technion_Israel">Technion_Israel</a></td> | ||
| + | <td>2015</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2015&group=Technion_Israel">Parts</a></td> | ||
| + | <td class="project">Be Bold: Hit baldness at its root</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2015.igem.org/Team:TJU">TJU</a></td> | ||
| + | <td>2015</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2015&group=TJU">Parts</a></td> | ||
| + | <td class="project">Power Consortia</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2014.igem.org/Team:Purdue">Purdue</a></td> | ||
| + | <td>2014</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2014&group=Purdue">Parts</a></td> | ||
| + | <td class="project">Minecrobe: Bacillus subtilis Production of Corn Phytosiderophores to Combat Malnutrition</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2014.igem.org/Team:LA%20Biohackers">LA Biohackers</a></td> | ||
| + | <td>2014</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2014&group=LA_Biohackers">Parts</a></td> | ||
| + | <td class="project">Boot up a Genome</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2014.igem.org/Team:Calgary">Calgary</a></td> | ||
| + | <td>2014</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2014&group=Calgary">Parts</a></td> | ||
| + | <td class="project">B.s. Detector ÔÇô A Multiplexed Diagnostic Device</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2014.igem.org/Team:LMU-Munich">LMU-Munich</a></td> | ||
| + | <td>2014</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2014&group=LMU-Munich">Parts</a></td> | ||
| + | <td class="project">ÔÇ×BaKillusÔÇť ÔÇô Engineering a pathogen-hunting microbe</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2014.igem.org/Team:Paris%20Bettencourt">Paris Bettencourt</a></td> | ||
| + | <td>2014</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2014&group=Paris_Bettencourt">Parts</a></td> | ||
| + | <td class="project">The smell of us</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2014.igem.org/Team:Toulouse">Toulouse</a></td> | ||
| + | <td>2014</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2014&group=Toulouse">Parts</a></td> | ||
| + | <td class="project">LetÔÇÖs save our trees with SubtiTree!</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2013.igem.org/Team:HZAU-China">HZAU-China</a></td> | ||
| + | <td>2013</td> | ||
| + | <td>Parts</td> | ||
| + | <td class="project">Safe moving vaccine factory</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2013.igem.org/Team:Groningen">Groningen</a></td> | ||
| + | <td>2013</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2013&group=Groningen">Parts</a></td> | ||
| + | <td class="project">Engineering Bacillus subtilis to self-assemble into a biofilm that coats medical implants with spider silk.</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2013.igem.org/Team:Hong_Kong_CUHK">Hong_Kong_CUHK</a></td> | ||
| + | <td>2013</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2013&group=Hong_Kong_CUHK">Parts</a></td> | ||
| + | <td class="project">Switch off PAHs</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2013.igem.org/Team:USTC_CHINA">USTC_CHINA</a></td> | ||
| + | <td>2013</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2013&group=USTC_CHINA">Parts</a></td> | ||
| + | <td class="project">T-VACCINE</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2013.igem.org/Team:Edinburgh">Edinburgh</a></td> | ||
| + | <td>2013</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2013&group=Edinburgh">Parts</a></td> | ||
| + | <td class="project">WastED</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2013.igem.org/Team:METU_Turkey">METU_Turkey</a></td> | ||
| + | <td>2013</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2013&group=METU_Turkey">Parts</a></td> | ||
| + | <td class="project">Bee subtilis</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2013.igem.org/Team:Newcastle">Newcastle</a></td> | ||
| + | <td>2013</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2013&group=Newcastle">Parts</a></td> | ||
| + | <td class="project">L-forms: Bacteria without a cell wall - a novel chassis for synthetic biology</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2013.igem.org/Team:UChicago">UChicago</a></td> | ||
| + | <td>2013</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2013&group=UChicago">Parts</a></td> | ||
| + | <td class="project">Keratinase Expression System in E. coli and B. subtilis</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2013.igem.org/Team:UNITN-Trento">UNITN-Trento</a></td> | ||
| + | <td>2013</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2013&group=UNITN-Trento">Parts</a></td> | ||
| + | <td class="project">B. fruity</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2012.igem.org/Team:UNAM_Genomics_Mexico">UNAM_Genomics_Mexico</a></td> | ||
| + | <td>2012</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2012&group=UNAM_Genomics_Mexico">Parts</a></td> | ||
| + | <td class="project">Bacillus booleanus</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2012.igem.org/Team:LMU-Munich">LMU-Munich</a></td> | ||
| + | <td>2012</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2012&group=LMU-Munich">Parts</a></td> | ||
| + | <td class="project">Beadzillus: Fundamental BioBricks for Bacillus subtilis and spores as a platform for protein display</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2012.igem.org/Team:Cambridge">Cambridge</a></td> | ||
| + | <td>2012</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2012&group=Cambridge">Parts</a></td> | ||
| + | <td class="project">Parts for a reliable and field ready biosensing platform</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2012.igem.org/Team:Groningen">Groningen</a></td> | ||
| + | <td>2012</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2012&group=Groningen">Parts</a></td> | ||
| + | <td class="project">The Food Warden. ItÔÇÖs rotten and you know it!</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2012.igem.org/Team:Warsaw">Warsaw</a></td> | ||
| + | <td>2012</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2012&group=Warsaw">Parts</a></td> | ||
| + | <td class="project">B. subtilis: supporting actor of the iGEM stage</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2012.igem.org/Team:HKUST-Hong_Kong">HKUST-Hong_Kong</a></td> | ||
| + | <td>2012</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2012&group=HKUST-Hong_Kong">Parts</a></td> | ||
| + | <td class="project">B. hercules---The Terminator of Colon Cancer</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2011.igem.org/Team:GeorgiaTech">GeorgiaTech</a></td> | ||
| + | <td>2011</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2011&group=GeorgiaTech">Parts</a></td> | ||
| + | <td class="project">De Novo Adaptation of Streptococcus thermophilus CRISPR1 Defense in Bacillus Subtilis</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2010.igem.org/Team:Newcastle">Newcastle</a></td> | ||
| + | <td>2010</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2010&group=Newcastle">Parts</a></td> | ||
| + | <td class="project">BacillaFilla: Filling Microcracks in Concrete</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2010.igem.org/Team:Groningen">Groningen</a></td> | ||
| + | <td>2010</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2010&group=Groningen">Parts</a></td> | ||
| + | <td class="project">Hydrophobofilm --- a self assembling hydrophobic biofilm</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2010.igem.org/Team:Imperial_College_London">Imperial_College_London</a></td> | ||
| + | <td>2010</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2010&group=Imperial_College_London">Parts</a></td> | ||
| + | <td class="project">Parasight ÔÇô Parasite detection with a rapid response</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2009.igem.org/Team:Cornell">Cornell</a></td> | ||
| + | <td>2009</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2009&group=Cornell">Parts</a></td> | ||
| + | <td class="project">Engineering the Bacillus Subtilis Metal Ion Homeostasis System to Serve as a Cadmium Responsive Biosensor</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2009.igem.org/Team:Newcastle">Newcastle</a></td> | ||
| + | <td>2009</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2009&group=Newcastle">Parts</a></td> | ||
| + | <td class="project">Bac-man: sequestering cadmium into Bacillus spores</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2008.igem.org/Team:Imperial_College">Imperial_College</a></td> | ||
| + | <td>2008</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Imperial_College">Parts</a></td> | ||
| + | <td class="project">Designer Genes ÔÇô Biofabricator subtilis</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2008.igem.org/Team:Cambridge">Cambridge</a></td> | ||
| + | <td>2008</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Cambridge">Parts</a></td> | ||
| + | <td class="project">Cambridge iBrain: Foundations for an Artificial Nervous System using Self-Organizing Electrical Patterning</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a target="_blank" href="http://2008.igem.org/Team:Newcastle_University">Newcastle_University</a></td> | ||
| + | <td>2008</td> | ||
| + | <td><a target="_blank" href="https://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Newcastle_University">Parts</a></td> | ||
| + | <td class="project">A Computational Intelligence Approach to Developing a Diagnostic Biosensor: The Newcastle BugBusters Project</td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | ==Plasmid backbone== | ||
| + | |||
| + | {{:Plasmid_backbones/Overview}} | ||
| + | |||
| + | |||
| + | |||
| + | The plasmids for ''B. Subtilis'' you can find in the registry are usually not built as standard registry plasmid so far. They come from third parties sources, that have been biobricked. Hence, these plasmid are often not silent. Cloning inside often works in different conditions than with standard E. coli plasmids. You may need to optimize again your ligation conditions. | ||
| + | |||
| + | As a particular drawback we noticed using replicative plasmids, is that they are often subject to recombination. We invite you to read in detail the experiment done and the peer review page before choosing a plasmid and a strategy. | ||
| + | |||
| + | There are 2 main kind of plasmids for B. Subtilis: | ||
| + | |||
| + | * Replicative or also called episomal plasmids, are the same kind of plasmid with a replication origin and a resistance gene we are used to work with in E. Coli. These plasmids are often made to be multihost, so that they can be cloned using E. Coli and then the miniprep can be transformed into ''B. subtilis'' by starvation or electroporation. (Note that some replicative plasmids requires to be transformed into RecA+ E. Coli strains first before incorporating by starvation) | ||
| + | |||
| + | * Integration plasmids is the most used technique by specialist in B. subtilis. They are E. Coli plasmids with two homologous sequence with two fragments of a chromosomal gene of B. Subtilis, and a resistance cassette inserted in between. If your brick is cloned in between these two homologous region, the vector act as a shuttle that integrate your construct into the chromosome. | ||
| + | |||
| + | |||
| + | Note that you should check your backbone and biobrick part sequence whether more than one side can be cut by same enzyme.You should not use if it has more than one same restriction enzyme site. | ||
| + | |||
| + | If you always see same bands in every gel images as different bp length from part page, you can try to transform your plasmid. Make sure that antibiotic you will use matches with which the backbone has.. | ||
| + | |||
| + | And also if you see another band too, always at the same place and since first run. You can try transform your plasmid and check the viability when you exclude the possibility of contamination or any other things which can cause this problem in your lab. Another possibility could be that the part itself has already additional contaminated DNAs. | ||
| + | |||
| + | |||
| + | <parttable>bsubtilis_plasmid_backbone</parttable> | ||
| + | |||
| + | ==Constitutive promoters== | ||
{{:Promoters/Catalog/B._subtilis/Constitutive}} | {{:Promoters/Catalog/B._subtilis/Constitutive}} | ||
| + | |||
| + | ==Positively regulated promoters== | ||
{{:Promoters/Catalog/B._subtilis/Positive}} | {{:Promoters/Catalog/B._subtilis/Positive}} | ||
| + | |||
| + | ==Repressible promoters== | ||
{{:Promoters/Catalog/B._subtilis/Repressible}} | {{:Promoters/Catalog/B._subtilis/Repressible}} | ||
| + | |||
| + | ==Ribosome binding sites== | ||
| + | |||
| + | ''B. subtilis'' have the same system of RBS than E. coli to drive its transcription. Hoever, the RBS sequences are not always compatible since ''B. subtilis'' ribsomomal RNA (rRNA) is slightly different in sequence compare to ''E. coli''. | ||
| + | |||
| + | In this list, you can find the most common RBSs usually use in B. subtilis. They are classified by strengh in a qualitative manner, since no strengh reference ranking have been established for ''B. subtilis'' yet. It is not obvious to transport directly the system for ''E. coli'' since we need to have a promoter that is exactly the same strengh as an E. coli one to make both scales comparable between the organism. | ||
| + | |||
| + | We can also notice that ''E. coli'' RBSs usually doesn't works for ''B. subtilis'', but it is not rare to find ''B. subtilis'' RBSs that works fine in ''E. coli'' (the SpoVg RBS for instance). | ||
| + | |||
| + | Please go in the description of each parts for more information. | ||
| + | |||
| + | <parttable>bsubtilis_rbs</parttable> | ||
| + | |||
| + | ==Protein coding sequences== | ||
| + | |||
| + | {{:Protein coding sequences/Overview}} | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | '''Note of the codon availability in B. subtilis:''' B. subtilis is more codon tolerent than ''E. coli'', which means that you can generally express any protein from ''E. coli'' into ''B. subtilis'' without any problem. However, some proteins are said to be for subtilis for two reasons: One is because they comes from B. subtilis genome, (which means that you may have trouble in expressing them in ''E. coli'') or because they are made to trigger a certain effect in ''B. subtilis'' (for instance kill it). | ||
| + | |||
| + | |||
| + | <parttable>bsubtilis_protein_coding_sequence</parttable> | ||
| + | |||
| + | <!-- To include a part in this table, include the categories "//chassis/prokaryote/bsubtilis" set the part type to Coding under the Hard Information tab of the part. --> | ||
| + | |||
| + | ==DNA parts for building integration vector== | ||
| + | |||
| + | Any ''E. coli'' plasmid can be converted into an integration vector if one add two pieces of a a gene sequence that is homologous to a gene into ''B. subtilis'' genes, and an antibiotic resistance cassette or other screening method. | ||
| + | <br /> | ||
| + | |||
| + | {{:DNA/Chromosomal_integration/{{PAGENAME}}/Overview}} | ||
| + | |||
| + | <parttable>bsubtilis_DNA</parttable> | ||
==References== | ==References== | ||
| Ôłĺ | {{: | + | {{:{{PAGENAME}}/References}} |
__NOTOC__ __NOEDITSECTION__ | __NOTOC__ __NOEDITSECTION__ | ||
Latest revision as of 15:52, 16 July 2020
< Back to Catalog
Bacillus subtilis is gram-positive model organism. Thus, much is known about this organism. The genome of Bacillus subtilis strain 168 has been sequenced.
Although the current part collection for B. subtilis is small, many are now using B. subtilis as a candidate host for synthetic devices and systems. Please read more about the advantages and disadvantages of using B. subtilis as a chassis.
| Plasmid backbones (?) | Promoters (?) | Ribosome binding sites (?) | |||
| Protein coding sequences (?) | DNA parts (?) |
| Help: Want to know more about Bacillus subtilis? See the help pages for more information. |
Previous iGEM Bacillus subtilis Projects
| Team | Year | Parts | Project Title |
|---|---|---|---|
| DTU-Denmark | 2015 | Parts | The Synthesizer: Development of antibiotic libraries through Multiplex Automated Genome Engineering |
| Groningen | 2015 | Parts | Blue Bio Energy |
| Nagahama | 2015 | Parts | ''ÚŽÖŔöÁň║ź''ŃÇÇFlavorator: New food preservation method by rose odor E. coli |
| Nanjing-China | 2015 | Parts | Metallosniper: ┬Śinnovative total solution for heavy metals |
| NEFU_China | 2015 | Parts | Yogurt Guarder |
| Stanford-Brown | 2015 | Parts | biOrigami: A New Approach to Reduce the Cost of Space Missions |
| Technion_Israel | 2015 | Parts | Be Bold: Hit baldness at its root |
| TJU | 2015 | Parts | Power Consortia |
| Purdue | 2014 | Parts | Minecrobe: Bacillus subtilis Production of Corn Phytosiderophores to Combat Malnutrition |
| LA Biohackers | 2014 | Parts | Boot up a Genome |
| Calgary | 2014 | Parts | B.s. Detector ÔÇô A Multiplexed Diagnostic Device |
| LMU-Munich | 2014 | Parts | ÔÇ×BaKillusÔÇť ÔÇô Engineering a pathogen-hunting microbe |
| Paris Bettencourt | 2014 | Parts | The smell of us |
| Toulouse | 2014 | Parts | LetÔÇÖs save our trees with SubtiTree! |
| HZAU-China | 2013 | Parts | Safe moving vaccine factory |
| Groningen | 2013 | Parts | Engineering Bacillus subtilis to self-assemble into a biofilm that coats medical implants with spider silk. |
| Hong_Kong_CUHK | 2013 | Parts | Switch off PAHs |
| USTC_CHINA | 2013 | Parts | T-VACCINE |
| Edinburgh | 2013 | Parts | WastED |
| METU_Turkey | 2013 | Parts | Bee subtilis |
| Newcastle | 2013 | Parts | L-forms: Bacteria without a cell wall - a novel chassis for synthetic biology |
| UChicago | 2013 | Parts | Keratinase Expression System in E. coli and B. subtilis |
| UNITN-Trento | 2013 | Parts | B. fruity |
| UNAM_Genomics_Mexico | 2012 | Parts | Bacillus booleanus |
| LMU-Munich | 2012 | Parts | Beadzillus: Fundamental BioBricks for Bacillus subtilis and spores as a platform for protein display |
| Cambridge | 2012 | Parts | Parts for a reliable and field ready biosensing platform |
| Groningen | 2012 | Parts | The Food Warden. ItÔÇÖs rotten and you know it! |
| Warsaw | 2012 | Parts | B. subtilis: supporting actor of the iGEM stage |
| HKUST-Hong_Kong | 2012 | Parts | B. hercules---The Terminator of Colon Cancer |
| GeorgiaTech | 2011 | Parts | De Novo Adaptation of Streptococcus thermophilus CRISPR1 Defense in Bacillus Subtilis |
| Newcastle | 2010 | Parts | BacillaFilla: Filling Microcracks in Concrete |
| Groningen | 2010 | Parts | Hydrophobofilm --- a self assembling hydrophobic biofilm |
| Imperial_College_London | 2010 | Parts | Parasight ÔÇô Parasite detection with a rapid response |
| Cornell | 2009 | Parts | Engineering the Bacillus Subtilis Metal Ion Homeostasis System to Serve as a Cadmium Responsive Biosensor |
| Newcastle | 2009 | Parts | Bac-man: sequestering cadmium into Bacillus spores |
| Imperial_College | 2008 | Parts | Designer Genes ÔÇô Biofabricator subtilis |
| Cambridge | 2008 | Parts | Cambridge iBrain: Foundations for an Artificial Nervous System using Self-Organizing Electrical Patterning |
| Newcastle_University | 2008 | Parts | A Computational Intelligence Approach to Developing a Diagnostic Biosensor: The Newcastle BugBusters Project |
Plasmid backbone
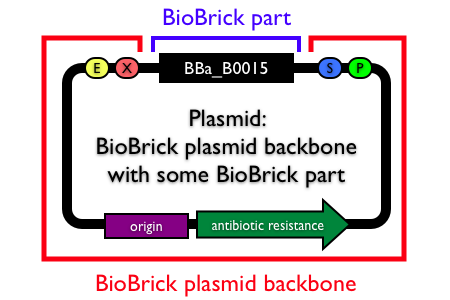
Plasmids are circular, double-stranded DNA molecules typically containing a few thousand base pairs that replicate within the cell independently of the chromosomal DNA. Plasmid DNA is easily purified from cells, manipulated using common lab techniques and incorporated into cells. Most BioBrick parts in the Registry are maintained and propagated on plasmids. Thus, construction of BioBrick parts, devices and systems usually requires working with plasmids.
Note: In the Registry, plasmids are made up of two distinct components:
- the BioBrick part, device or system that is located in the BioBrick cloning site, between (and excluding) the BioBrick prefix and suffix.
- the plasmid backbone which propagates the BioBrick part. The plasmid backbone is defined as the sequence beginning with the BioBrick suffix, including the replication origin and antibiotic resistance marker, and ending with the BioBrick prefix. [Note that the plasmid backbone itself can be composed of BioBrick parts.]
Many BioBrick parts in the Registry are maintained on more than one plasmid backbone!
The plasmids for B. Subtilis you can find in the registry are usually not built as standard registry plasmid so far. They come from third parties sources, that have been biobricked. Hence, these plasmid are often not silent. Cloning inside often works in different conditions than with standard E. coli plasmids. You may need to optimize again your ligation conditions.
As a particular drawback we noticed using replicative plasmids, is that they are often subject to recombination. We invite you to read in detail the experiment done and the peer review page before choosing a plasmid and a strategy.
There are 2 main kind of plasmids for B. Subtilis:
- Replicative or also called episomal plasmids, are the same kind of plasmid with a replication origin and a resistance gene we are used to work with in E. Coli. These plasmids are often made to be multihost, so that they can be cloned using E. Coli and then the miniprep can be transformed into B. subtilis by starvation or electroporation. (Note that some replicative plasmids requires to be transformed into RecA+ E. Coli strains first before incorporating by starvation)
- Integration plasmids is the most used technique by specialist in B. subtilis. They are E. Coli plasmids with two homologous sequence with two fragments of a chromosomal gene of B. Subtilis, and a resistance cassette inserted in between. If your brick is cloned in between these two homologous region, the vector act as a shuttle that integrate your construct into the chromosome.
Note that you should check your backbone and biobrick part sequence whether more than one side can be cut by same enzyme.You should not use if it has more than one same restriction enzyme site.
If you always see same bands in every gel images as different bp length from part page, you can try to transform your plasmid. Make sure that antibiotic you will use matches with which the backbone has..
And also if you see another band too, always at the same place and since first run. You can try transform your plasmid and check the viability when you exclude the possibility of contamination or any other things which can cause this problem in your lab. Another possibility could be that the part itself has already additional contaminated DNAs.
| Name | Description | Resistance | Replicon | Copy number | Chassis | Length |
|---|---|---|---|---|---|---|
| BBa_I742123 | Multi-host vector pTG262 converted to BioBrick vector | C | Multi-host | Multi-host | 5564 | |
| BBa_J179000 | pBS1C: Bacillus subtilis vector, amyE integration, CAM-resistance | A(E. coli) C(B. subtilis) | B. subtilis + E. coli | 6105 | ||
| BBa_J179001 | pBS2E: Bacillus subtilis vector, lacA integration, MLS resistance | A(E. coli) E(B. subtilis) | B. subtilis + E. coli | 6258 | ||
| BBa_J179002 | pBs4S: Bacillus subtilis vector, thrC integration, spec resistance | A(E. coli) S(B. subtilis) | pDG1731 | integrative in B. subtilis | B. subtilis + E. coli | 4573 |
| BBa_K090402 | B. subtilis Episomal Vector with Constitutive GFP | 6337 | ||||
| BBa_K090403 | Gram-positive Shuttle Vector for Chromosomal Integration | 5632 | ||||
| BBa_K1065204 | Efe+Bba_B0015 in BBa_K823024 (pXyl) | A(E. coli) S(B. subtilis) | B. subtilis + E. coli | 6017 | ||
| BBa_K1085014 | pSB1AC3-HySp: Integrational backbone into B. subtilis amyE locus with IPTG inducible promoter | 7625 | ||||
| BBa_K1185004 | Integration vector for Bacillus subtilis derived from pSac-Cm | AC | multi-host | multi-host | 5210 | |
| BBa_K1364021 | Integrative plasmid for Bacillus subtilis (pSBbs4E) | 5760 | ||||
| BBa_K818000 | Integration vector for Bacillus subtilis derived from pSac-Cm | AC | Multi host | Multi host | 5223 | |
| BBa_K823021 | pSBBs1C-lacZ (lacZ reporter vector for B. subtilis) | A(E. coli) C(B. subtilis) | pAC6 | integrative in B. subtilis | E.coli and B. subtilis | 9792 |
| BBa_K823022 | pSBBs4S: Empty backbone for integration into Bacillus subtilis thrC locus | A(E. coli) S(B. subtilis) | pDG1731 | integrative in B. subtilis | B. subtilis + E. coli | 4573 |
| BBa_K823023 | pSBBs1C: Empty backbone for integration into Bacillus subtilis amyE locus | A(E. coli) C(B. subtilis) | B. subtilis + E. coli | 6105 | ||
| BBa_K823024 | pSBBs4S-Pxyl: Integrative expression vector for Bacillus subtilis | A(E. coli) S(B. subtilis) | B. subtilis + E. coli | 4794 | ||
| BBa_K823025 | pSBBs3C-luxABCDE (lux reporter vector for B.subtilis) | A(E. coli) + C(B. subtilis) | pAH328 | integrative in B. subtilis | B. subtilis + E. coli | 10640 |
| BBa_K823027 | pSBBs2E: Empty backbone for integration into Bacillus subtilis lacA locus | A(E. coli) E(B. subtilis) | B. subtilis + E. coli | 6193 |
Constitutive promoters
The promoters here are B. subtilis promoters that are constitutive meaning that the activity of these promoters should only be regulated by the levels of RNA polymerase and the appropriate σ factor.
The sequence of these promoter are adapted to the σ factor of B. subtilis. However, some of these promoter also works in E. coli. Generally speaking, standard E. coli promoters don't work (or are very weak) in B. subtilis strains, whereas the contrary generally works. However, it doesn't mean that the efficiency will be the same in both strains. The pVeg promoter, for instance, works fine at a high level of expression in both E. coli and B. subtilis strains - Contribution from User: Cyrpaut (31 October 2011)
Constitutive B. subtilis σA promoters
This section lists promoters that are recognized by B. subtilis σA RNAP. σA is the major B. subtilis sigma factor so there should be RNAP present to transcribe these promoters under most growth conditions (although maximally during exponential growth).
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_K143012 | Promoter veg a constitutive promoter for B. subtilis | . . . aaaaatgggctcgtgttgtacaataaatgt | 97 | 6561 | In stock | ||
| BBa_K143013 | Promoter 43 a constitutive promoter for B. subtilis | . . . aaaaaaagcgcgcgattatgtaaaatataa | 56 | 11873 | Not in stock | ||
| BBa_K780003 | Strong constitutive promoter for Bacillus subtilis | . . . aattgcagtaggcatgacaaaatggactca | 36 | 1559 | It's complicated | ||
| BBa_K823000 | PliaG | . . . caagcttttcctttataatagaatgaatga | 121 | 7765 | In stock | ||
| BBa_K823002 | PlepA | . . . tctaagctagtgtattttgcgtttaatagt | 157 | 7245 | In stock | ||
| BBa_K823003 | Pveg | . . . aatgggctcgtgttgtacaataaatgtagt | 237 | 14126 | In stock |
Constitutive B. subtilis σB promoters
This section lists promoters that are recognized by B. subtilis σB RNAP. σB is the major stationary phase E. coli sigma factor. Use these promoters when you want high promoter activity during stationary phase or during starvation.
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_K143010 | Promoter ctc for B. subtilis | . . . atccttatcgttatgggtattgtttgtaat | 56 | 4297 | Not in stock | ||
| BBa_K143011 | Promoter gsiB for B. subtilis | . . . taaaagaattgtgagcgggaatacaacaac | 38 | 2765 | Not in stock | ||
| BBa_K143013 | Promoter 43 a constitutive promoter for B. subtilis | . . . aaaaaaagcgcgcgattatgtaaaatataa | 56 | 11873 | Not in stock |
Positively regulated promoters
The B. subtilis promoters of this section is the ones that are said to be positively regulated. It means that meaning their expression level increase with the help of another third party protein called transcription activator (This category exclude the sigma factor protein itself). With the appropriate protein, you would be able to increase the activity of your promoter. Please read the description and characterization of each parts for more details.
Positively regulated B. subtilis σA promoters
This section lists the promoters recognized by B. subtilis σA RNA polymerase sub-unit. σA is the major B. subtilis sigma factor that is present under most growth conditions (but maximal during exponential growth phase).
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_K090504 | Gram-Positive Strong Constitutive Promoter | . . . acatgggaaaactgtatgtatttgatcctc | 239 | 2275 | It's complicated |
Positively regulated B. subtilis σB promoters
This section lists promoters that are recognized by B. subtilis σB RNA polymerase. σB is the polymerase subunit that is the most present during the stationary growth phase. You can use these promoters if you want your construct to be mostly expressed during stationary growth phase or under starvation conditions.
There are no parts for this table
Repressible promoters
The B. subtilis promoters of this section are said negativly regulated promoters, because they can be repressed by the expression of a third party protein. The inhibition can be released by the addition of a molecule, like for the LacI E. coli promoter.
In the following biobricks, the proposed promoters are build with the fusion of one or several operons with a σA type contitutive promoter.
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_K090501 | Gram-Positive IPTG-Inducible Promoter | . . . tggaattgtgagcggataacaattaagctt | 107 | 2054 | It's complicated | ||
| BBa_K143014 | Promoter Xyl for B.subtilis | . . . agtttgtttaaacaacaaactaataggtga | 82 | 2112 | Not in stock | ||
| BBa_K143015 | Promoter hyper-spank for B. subtilis | . . . aatgtgtgtaattgtgagcggataacaatt | 101 | 2527 | Not in stock |
In the future, we may also find promoters builded with the σB promoter.
There are no parts for this table
Ribosome binding sites
B. subtilis have the same system of RBS than E. coli to drive its transcription. Hoever, the RBS sequences are not always compatible since B. subtilis ribsomomal RNA (rRNA) is slightly different in sequence compare to E. coli.
In this list, you can find the most common RBSs usually use in B. subtilis. They are classified by strengh in a qualitative manner, since no strengh reference ranking have been established for B. subtilis yet. It is not obvious to transport directly the system for E. coli since we need to have a promoter that is exactly the same strengh as an E. coli one to make both scales comparable between the organism.
We can also notice that E. coli RBSs usually doesn't works for B. subtilis, but it is not rare to find B. subtilis RBSs that works fine in E. coli (the SpoVg RBS for instance).
Please go in the description of each parts for more information.
| Name | Sequence | Description | Relative Strength | Predicted Strength |
|---|---|---|---|---|
| BBa_K090505 | aaaggaggtgt | ''Bacillus subtilis'' consensus RBS | ||
| BBa_K090506 | agaggtggtgt | ''Bacillus subtilis'' weak RBS | ||
| BBa_K104003 | agggggccg | RBS_spaR | ||
| BBa_K104005 | ggggcgttg | RBS_spaK | ||
| BBa_K104009 | agaggagg | RBS_gfp | ||
| BBa_K1351028 | aaggagggata | LMU Bacillus RBS collection 1 | ||
| BBa_K1351029 | agaggaggata | LMU Bacillus RBS collection 2 | ||
| BBa_K1351030 | aaggagagata | LMU Bacillus RBS collection 3 | ||
| BBa_K1351031 | aggagaggata | LMU Bacillus RBS collection 4 | ||
| BBa_K1351032 | aagaggagata | LMU Bacillus RBS collection 5 | ||
| BBa_K1351033 | agaaagggata | LMU Bacillus RBS collection 6 | ||
| BBa_K1351034 | aagaagagata | LMU Bacillus RBS collection 7 | ||
| BBa_K143020 | taaaggaggaa | GsiB ribosome binding site (RBS) for B. subtilis | ||
| BBa_K143021 | aaaggtggtgaa | SpoVG ribosome binding site (RBS) for B. subtilis | ||
| BBa_K2586010 | . . . tgctacaattactagaggtttaaaggagga | Palf4-RBS: Palf4 linked to a RBS | ||
| BBa_K3831005 | gtaataagtaggttaggagagg | Synthetic RBS_a | ||
| BBa_K3831006 | . . . actacacgacaattaaagaaggtatttttt | Synthetic RBS_b | ||
| BBa_K3831007 | ggtggaaaggaggtgatcgac | Native RBS R2 | ||
| BBa_K3831008 | ggtgggaaggagggggttcgac | RBS R6 | ||
| BBa_K3831009 | gggatagacccagggggaggttttttt | Synthetic RBS_c | ||
| BBa_K3831014 | gctcttaaggaggattttaga | Native RBS R1 | ||
| BBa_K3831018 | gattaactaataaggaggacaaac | Native RBS R0 | ||
| BBa_K3831020 | ggtgggaaggaggacattcgac | Artificial RBS R3 | ||
| BBa_K3831047 | aaaaaaacctccccctgggtctatccc | Synthetic RBS_c - reversed | ||
| BBa_K3831049 | gtcgaaccccctccttcccacc | RBS R6 - reversed | ||
| BBa_K3831051 | gtcgatcacctcctttccacc | RBS R2 - reversed | ||
| BBa_K3831053 | . . . aaataccttctttaattgtcgtgtagtaat | Synthetic RBS_b - reversed | ||
| BBa_K3831055 | cctctcctaacctacttattac | Synthetic RBS_a - reversed | ||
| BBa_K4382009 | aaggaggaaggatca | RBS for Bacillus Subtilis | ||
| BBa_K780001 | atattaagaggaggag | Strong RBS for Bacillus Subtilis | ||
| BBa_K780002 | agagaacaaggagggg | Strong RBS for Bacillus Subtilis |
Protein coding sequences
Protein coding sequences are DNA sequences that are transcribed into mRNA and in which the corresponding mRNA molecules are translated into a polypeptide chain. Every three nucleotides, termed a codon, in a protein coding sequence encodes 1 amino acid in the polypeptide chain. In some cases, different chassis may either map a given codon to a different sequence or may use different codons more or less frequently. Therefore some protein coding sequences may be optimized for use in a particular chassis.
In the Registry, protein coding sequences begin with a start codon (usually ATG) and end with a stop codon (usually with a double stop codon TAA TAA). Protein coding sequences are often abbreviated with the acronym CDS.
Although protein coding sequences are often considered to be basic parts, in fact proteins coding sequences can themselves be composed of one or more regions, called protein domains. Thus, a protein coding sequence could either be entered as a basic part or as a composite part of two or more protein domains.
- The N-terminal domain of a protein coding sequence is special in a number of ways. First, it always contains a start codon, spaced at an appropriate distance from a ribosomal binding site. Second, many coding regions have special features at the N terminus, such as protein export tags and lipoprotein cleavage and attachment tags. These occur at the beginning of a coding region, and therefore are termed Head domains.
- A protein domain is a sequence of amino acids which fold relatively independently and which are evolutionarily shuffled as a unit among different protein coding regions. The DNA sequence of such domains must maintain in-frame translation, and thus is a multiple of three bases. Since these protein domains are within a protein coding sequence, they are called Internal domains. Certain Internal domains have particular functions in protein cleavage or splicing and are termed Special Internal domains.
- Similarly, the C-terminal domain of a protein is special, containing at least a stop codon. Other special features, such as degradation tags, are also required to be at the extreme C-terminus. Again, these domains cannot function when internal to a coding region, and are termed Tail domains.
For more details on protein domains including how to assemble protein domains into protein coding sequences, please see Protein domains.
Protein coding sequences should be as follows
GAATTC GCGGCCGC T TCTAG [ATG ... TAA TAA] T ACTAGT A GCGGCCG CTGCAG
Note of the codon availability in B. subtilis: B. subtilis is more codon tolerent than E. coli, which means that you can generally express any protein from E. coli into B. subtilis without any problem. However, some proteins are said to be for subtilis for two reasons: One is because they comes from B. subtilis genome, (which means that you may have trouble in expressing them in E. coli) or because they are made to trigger a certain effect in B. subtilis (for instance kill it).
| Name | Protein | Description | Tag | Direction | UniProt | KEGG | Length | Status |
|---|---|---|---|---|---|---|---|---|
| BBa_E0040 | GFPmut3b | green fluorescent protein derived from jellyfish Aequeora victoria wild-type GFP (SwissProt: P42212 | None | Forward | 720 | In stock | ||
| BBa_E1010 | mRFP1 | **highly** engineered mutant of red fluorescent protein from Discosoma striata (coral) | None | Forward | 706 | In stock | ||
| BBa_K104004 | spaR CDS (Coding sequence) | 663 | Not in stock | |||||
| BBa_K104006 | spaK CDS (Coding Sequence) | 1182 | Not in stock | |||||
| BBa_K1085000 | RBS Start-codon Strep-tag SpiderSilkSubunitE1 | 268 | In stock | |||||
| BBa_K1085001 | RBS Start-codon SpiderSilkSubunitE1 | 226 | In stock | |||||
| BBa_K1085002 | SpiderSilkSubunitE2 | 210 | In stock | |||||
| BBa_K1085003 | SpiderSilkSubunitE2 Strep-tag | 252 | In stock | |||||
| BBa_K1085004 | RBS Start-codon SpiderSilkSubunitN1 Strep-tag | 199 | In stock | |||||
| BBa_K1085005 | RBS Start-codon SpiderSilkSubunitN1 | 157 | In stock | |||||
| BBa_K1085006 | SpiderSilkSubunitN2 | 129 | In stock | |||||
| BBa_K1085007 | SpiderSilkSubunitN2 Strep-tag | 171 | In stock | |||||
| BBa_K1085010 | RBS Start-codon EstA Strep-tag SpiderSilkSubunitE1 | 907 | It's complicated | |||||
| BBa_K1085012 | RBS Start-codon EstA Strep-tag SpiderSilkSubunitN1 | 838 | In stock | |||||
| BBa_K1085029 | FliZ Strep-tag SpiderSilkSubunitE1 | 339 | In stock | |||||
| BBa_K1085034 | MotB Strep-tag SilkSubunitN1 | 309 | In stock | |||||
| BBa_K1085050 | des knock out | 3604 | Not in stock | |||||
| BBa_K1122000 | SinR transcription factor | 333 | It's complicated | |||||
| BBa_K1122666 | Ferric uptake repressor | 450 | It's complicated | |||||
| BBa_K1351000 | beta-Neurexin-derived peptide binding to S. aureus adhesin SdrC | 33 | In stock | |||||
| BBa_K1351001 | Contactin-4-derived peptide binding to NADH Oxidase of S. pneumoniae | 42 | In stock | |||||
| BBa_K1351002 | Chondroitin 4 sulfotransferase-derived peptide binding to NADH Oxidase of S. pneumoniae | 39 | In stock | |||||
| BBa_K1351003 | Laminin-derived peptide binding to NADH Oxidase of S. pneumoniae | 27 | In stock | |||||
| BBa_K1351006 | CWB domain of B. subtilis major autolysin LytC | 999 | In stock | |||||
| BBa_K1351007 | CWB domain of B. subtilis minor autolysin LytE | 621 | In stock | |||||
| BBa_K1351008 | Signal peptide of B. subtilis endonuclease YhcR | 138 | In stock | |||||
| BBa_K1351010 | Cell wall-anchoring domain of B. subtilis endonuclease YhcR | 402 | In stock | |||||
| BBa_K1351011 | Bacillus subtilis sortase YhcS | 597 | In stock | |||||
| BBa_K1351012 | spaS Subtilin (antimicrobial peptide, Freiburg standard) | 168 | In stock | |||||
| BBa_K1351013 | spaS Subtilin (antimicrobial peptide) | 171 | In stock | |||||
| BBa_K1351017 | SdpI with RBS: Immunity against the cannibalsim toxin sdpC of B. subtilis | 333 | In stock | |||||
| BBa_K1351021 | Monomeric Red Fluorescent Protein from Discosoma striata | 675 | In stock | |||||
| BBa_K1351026 | Signal peptide of B. subtilis major autolysin LytC | 75 | In stock | |||||
| BBa_K1351027 | Signal peptide of B. subtilis minor autolysin LytE | 75 | In stock | |||||
| BBa_K1362461 | mRFP1 | **highly** engineered mutant of red fluorescent protein from Discosoma striata (coral) with barcode | None | Forward | 706 | Not in stock | ||
| BBa_K1399000 | mRFP1 | RFP from Discosoma striata (coral) with AAV-ssrA degradation tag | SsrA-AAV | Forward | 714 | In stock | ||
| BBa_K1399001 | mRFP1 | RFP from Discosoma striata (coral) with LVA-ssrA degradation tag | SsrA-LVA degradation tag | Forward | 714 | In stock | ||
| BBa_K1399002 | mRFP1 | RFP from Discosoma striata (coral) with LAA-ssrA degradation tag (wt) | SsrA-LAA degradation tag | Forward | 714 | In stock | ||
| BBa_K1399003 | mRFP1 | RFP from Discosoma striata (coral) with DAS-ssrA degradation tag | SsrA-DAS degradation tag | Forward | 714 | In stock | ||
| BBa_K1399004 | GFPmut3b | GFP (mut3b) with LVA-ssrA degradation tag | SsrA-LVA degradation tag | Forward | 753 | In stock | ||
| BBa_K1399005 | GFPmut3b | GFP (mut3b) with AAV-ssrA degradation tag | SsrA-AAV degradation tag | Forward | 753 | In stock | ||
| BBa_K1399006 | GFPmut3b | GFP (mut3b) with LAA-ssrA degradation tag | SsrA-LAA degradation tag | Forward | 753 | In stock | ||
| BBa_K1399007 | GFPmut3b | GFP (mut3b) with SsrA-DAS+2 degradation tag | SsrA-DAS+2 degradation tag | Forward | 759 | In stock | ||
| BBa_K1399008 | GFPmut3b | GFP (mut3b) with DAS-ssrA degradation tag | SsrA-DAS degradation tag | Forward | 753 | In stock | ||
| BBa_K143032 | EpsE Molecular Clutch Gene of B. subtilis | 840 | In stock | |||||
| BBa_K143034 | LipA-EAK16-II Fusion Protein | 156 | It's complicated | |||||
| BBa_K143035 | LipA-Human Elastin (EP20-24-24) Fusion Protein | 705 | It's complicated | |||||
| BBa_K143036 | Xylose operon regulatory protein | 1056 | It's complicated | |||||
| BBa_K143037 | YtvA Blue Light Receptor for B.subtilis | 789 | It's complicated | |||||
| BBa_K143038 | SacB-EAK16-II Fusion Protein | 150 | It's complicated | |||||
| BBa_K143039 | SacB-Human Elastin (EP20-24-24) Fusion Protein | 699 | It's complicated | |||||
| BBa_K143063 | Xylose operon repressor protein - Terminator | 1193 | Not in stock | |||||
| BBa_K1701000 | GolB | 195 | In stock | |||||
| BBa_K1701001 | PbrR | 438 | It's complicated | |||||
| BBa_K1701004 | SUP | 267 | It's complicated | |||||
| BBa_K1701005 | TasA | 786 | It's complicated | |||||
| BBa_K1701006 | CotC | 201 | It's complicated | |||||
| BBa_K174012 | sleB, Bacillus subtilis germination gene with RBS | 932 | It's complicated | |||||
| BBa_K2057002 | Alginate lyase from Bacillus thuringiensis | No part sequence | ||||||
| BBa_K2273004 | Epr signal peptide of B. subtilis minor extracellular serine protease | 81 | It's complicated | |||||
| BBa_K2273005 | FliL signal peptide of B. subtilis flagellar protein | 69 | It's complicated | |||||
| BBa_K2273006 | FliZ signal peptide of B. subtilis flagellar biosynthetic protein | 81 | It's complicated | |||||
| BBa_K2273007 | GlpQ signal peptide of B. subtilis glycerolphosphate diester phosphodiesterase | 78 | It's complicated | |||||
| BBa_K2273008 | LipA signal peptide of B. subtilis lipoic acid synthase | 96 | It's complicated | |||||
| BBa_K2273009 | LytB signal peptide of B. subtilis modifier protein of major autolysin LytC | 75 | It's complicated | |||||
| BBa_K2273010 | LytC signal peptide of B. subtilis N-acetylmuramoyl-L-alanine amidase | 72 | It's complicated | |||||
| BBa_K2273011 | LytD signal peptide of B. subtilis peptidoglycan N-acetylglucosaminidase | 81 | It's complicated | |||||
| BBa_K2273015 | Minimal SpyCatcher codon adapted for Bacillus subtilis | 249 | It's complicated | |||||
| BBa_K2273023 | AmyE signal peptide of B. subtilis alpha-amylase | 99 | It's complicated | |||||
| BBa_K2273024 | AspB signal peptide of B. subtilis aspartate transaminase | 66 | It's complicated | |||||
| BBa_K2273025 | BglS signal peptide of B. subtilis endo-beta-1,3-1,4 glucanase | 84 | It's complicated | |||||
| BBa_K2273026 | CccA signal peptide of B. subtilis cytochrome c550 | 81 | It's complicated | |||||
| BBa_K2273027 | Bpr signal peptide of B. subtilis bacillopeptidase F | 90 | It's complicated | |||||
| BBa_K2273028 | Csn signal peptide of B. subtilis chitosanase | 105 | It's complicated | |||||
| BBa_K2273029 | DacB signal peptide of B. subtilis sporulation-specific penicillin-binding protein | 81 | It's complicated | |||||
| BBa_K2273030 | DacF signal peptide of B. subtilis penicillin-binding protein I | 69 | It's complicated | |||||
| BBa_K2273031 | DltD signal peptide of B. subtilis Protein DltD | 63 | It's complicated | |||||
| BBa_K2273032 | CitH signal peptide of B. subtilis malate dehydrogenase | 96 | It's complicated | |||||
| BBa_K2273043 | LytR signal peptide of B. subtilis phosphotransferase | 117 | It's complicated | |||||
| BBa_K2273051 | PbpX signal peptide of B. subtilis penicillin-binding protein X | 117 | It's complicated | |||||
| BBa_K2273052 | Pel signal peptide of B. subtilis pectate lyase C | 63 | It's complicated | |||||
| BBa_K2273053 | PelB signal peptide of B. subtilis pectate lyase | 72 | It's complicated | |||||
| BBa_K2273054 | PenP signal peptide of B. subtilis beta-lactamase | 108 | It's complicated | |||||
| BBa_K2273055 | PhoA signal peptide of B. subtilis alkaline phosphatase A | 93 | It's complicated | |||||
| BBa_K2273056 | PhoB signal peptide of B. subtilis alkaline phosphatase III | 96 | It's complicated | |||||
| BBa_K2273057 | PhrA signal peptide of B. subtilis Phosphatase RapA inhibitor | 75 | It's complicated | |||||
| BBa_K2273058 | PhrC signal peptide of B. subtilis Phosphatase RapC inhibitor | 81 | It's complicated | |||||
| BBa_K2273059 | PhrF signal peptide of B. subtilis Phosphatase RapF inhibitor | 72 | It's complicated | |||||
| BBa_K2273060 | PhrG signal peptide of B. subtilis Phosphatase RapG inhibitor | 63 | It's complicated | |||||
| BBa_K2273061 | PhrK signal peptide of B. subtilis Phosphatase RapK inhibitor | 57 | It's complicated | |||||
| BBa_K2273062 | RpmGB signal peptide of B. subtilis 50S ribosomal protein L33 2 | 78 | It's complicated | |||||
| BBa_K2273063 | SacB signal peptide of B. subtilis levansucrase | 87 | It's complicated | |||||
| BBa_K2273064 | SacC signal peptide of B. subtilis levanase | 72 | It's complicated | |||||
| BBa_K2273065 | SleB signal peptide of B. subtilis spore cortex-lytic enzyme | 93 | It's complicated | |||||
| BBa_K2273066 | SpoIID signal peptide of B. subtilis lytic transglycosylase | 102 | It's complicated | |||||
| BBa_K2273067 | SpoIIP signal peptide of B. subtilis cell wall hydrolase | 117 | It's complicated | |||||
| BBa_K2273068 | SpoIIQ signal peptide of B. subtilis stage II sporulation protein Q | 120 | It's complicated | |||||
| BBa_K2273069 | SpoIIR signal peptide of B. subtilis stage II sporulation protein R | 60 | It's complicated | |||||
| BBa_K2273070 | TyrA signal peptide of B. subtilis prephenate dehydrogenase | 69 | It's complicated | |||||
| BBa_K2273071 | Vpr signal peptide of B. subtilis minor extracellular serine protease | 93 | It's complicated | |||||
| BBa_K2273072 | WapA signal peptide of B. subtilis cell wall-associated protein precursor | 96 | It's complicated | |||||
| BBa_K2273073 | YbbC signal peptide of B. subtilis unknown product | 69 | It's complicated | |||||
| BBa_K2273074 | YbbE signal peptide of B. subtilis N-acetylmuramyl-L-alanine amidase | 75 | It's complicated | |||||
| BBa_K2273075 | YbbR signal peptide of B. subtilis CdaA regulatory protein CdaR | 84 | It's complicated | |||||
| BBa_K2273076 | YbdG signal peptide of B. subtilis Uncharacterized protein YbdG | 75 | It's complicated | |||||
| BBa_K2273077 | YbdN signal peptide of B. subtilis Uncharacterized protein YbdN | 75 | It's complicated | |||||
| BBa_K2273078 | YbfO signal peptide of B. subtilis Putative hydrolase YbfO | 84 | It's complicated | |||||
| BBa_K2273079 | YbxI signal peptide of B. subtilis Probable beta-lactamase YbxI | 69 | It's complicated | |||||
| BBa_K2273080 | YdbK signal peptide of B. subtilis Uncharacterized membrane protein YdbK | 99 | It's complicated | |||||
| BBa_K2273081 | YdhT signal peptide of B. subtilis Mannan endo-1,4-beta-mannosidase | 63 | It's complicated | |||||
| BBa_K2273092 | YjcN signal peptide of B. subtilis uncharacterized protein YjcN | 102 | It's complicated | |||||
| BBa_K2273093 | YjdB signal peptide of B. subtilis uncharacterized protein YjdB | 78 | It's complicated | |||||
| BBa_K2273094 | YjfA signal peptide of B. subtilis uncharacterized protein YjfA | 84 | It's complicated | |||||
| BBa_K2273095 | YjiA signal peptide of B. subtilis uncharacterized protein YjiA | 78 | It's complicated | |||||
| BBa_K2273096 | YkoJ signal peptide of B. subtilis uncharacterized protein YkoJ | 78 | It's complicated | |||||
| BBa_K2273097 | YkvV signal peptide of B. subtilis sporulation thiol-disulfide oxidoreductase A | 78 | It's complicated | |||||
| BBa_K2273098 | YkwD signal peptide of B. subtilis uncharacterized protein YkwD | 78 | It's complicated | |||||
| BBa_K2273099 | YlaE signal peptide of B. subtilis uncharacterized protein YlaE | 93 | It's complicated | |||||
| BBa_K2273100 | YlbL signal peptide of B. subtilis uncharacterized protein YlbL | 105 | It's complicated | |||||
| BBa_K2273101 | YlqB signal peptide of B. subtilis inhibitor of entry into sporulation | 81 | It's complicated | |||||
| BBa_K2273102 | YlxF signal peptide of B. subtilis FlaA locus 22.9 kDa protein | 111 | It's complicated | |||||
| BBa_K2273103 | AmyEEV version of B. subtilis alpha-amylase without its native signal peptide | 1881 | It's complicated | |||||
| BBa_K2273104 | AmyERFC25 version of B. subtilis alpha-amylase without its native signal peptide | 1881 | It's complicated | |||||
| BBa_K2586001 | GltT: glutamate and glyphosate transporter in B. subtilis | 1290 | It's complicated | |||||
| BBa_K2586002 | GltP: glutamate and glyphosate uptake transporter in Bacillus subtilis | 1245 | It's complicated | |||||
| BBa_K2586003 | aroE: 3-phosphoshikimate 1-carboxyvinyltransferase | 1287 | It's complicated | |||||
| BBa_K2586007 | aroA: 3-phosphoshikimate 1-carboxyvinyltransferase | 1284 | It's complicated | |||||
| BBa_K2586019 | GAT: Glyphosate N-Acetyltransferase | 440 | It's complicated | |||||
| BBa_K2660000 | Penetratin, cell-penetrating peptide | 51 | Not in stock | |||||
| BBa_K2660003 | mCherry codon optimized for Lactococcus lactis | 724 | Not in stock | |||||
| BBa_K2660006 | N-Cas9 | 3459 | It's complicated | |||||
| BBa_K2660007 | C-Cas9 | 648 | It's complicated | |||||
| BBa_K2660008 | nMag | 450 | It's complicated | |||||
| BBa_K2660009 | pMag | 450 | It's complicated | |||||
| BBa_K294055 | GFPmut3b | GFP RFP Hybrid | None | 720 | In stock | |||
| BBa_K302001 | yneA coding sequence | 321 | Not in stock | |||||
| BBa_K302006 | spaI | 498 | Not in stock | |||||
| BBa_K302007 | spaF | 744 | Not in stock | |||||
| BBa_K302008 | spaE | 756 | Not in stock | |||||
| BBa_K302010 | sfp | 678 | Not in stock | |||||
| BBa_K302011 | swrA | 432 | Not in stock | |||||
| BBa_K302033 | mazF | 339 | In stock | |||||
| BBa_K3128008 | mRFP1 | Engineered mutant of red fluorescent protein with RFC21 restriction sites | None | Forward | 690 | Not in stock | ||
| BBa_K3590005 | SacB_v1 | 87 | It's complicated | |||||
| BBa_K3590006 | SacB signal peptide of Bacillus subtilis levansucrase version 2 | 87 | Not in stock | |||||
| BBa_K3590013 | Cellulose binding module | 495 | Not in stock | |||||
| BBa_K3590014 | Cohesin derived from Acetivibrio cellulolyticus | 438 | Not in stock | |||||
| BBa_K3590015 | Dockerin derived from Acetivibrio cellulolyticus | 228 | Not in stock | |||||
| BBa_K3590016 | Cohesin derived from Bacteroides cellulosolvens | 450 | Not in stock | |||||
| BBa_K3590017 | Dockerin derived from Bacteroides cellulosolvens | 243 | Not in stock | |||||
| BBa_K3590018 | Cohesin derived from Clostridium thermocellum | 435 | Not in stock | |||||
| BBa_K3590019 | Dockerin derived from Clostridium thermocellum | 219 | Not in stock | |||||
| BBa_K3590020 | 3x LysM domain derived from Bacillus subtilis LytE version 1 | 621 | Not in stock | |||||
| BBa_K3590021 | 3x LysM domain derived from Bacillus subtilis LytE version 2 | 621 | Not in stock | |||||
| BBa_K3590022 | Lysozyme derived from Bacillus licheniformis | 951 | Not in stock | |||||
| BBa_K3590024 | Microvirin | 324 | Not in stock | |||||
| BBa_K3590025 | MlrA | 1008 | Not in stock | |||||
| BBa_K3590026 | MlrB | 1623 | Not in stock | |||||
| BBa_K3590027 | MlrC | 1584 | Not in stock | |||||
| BBa_K3590028 | Linker 1 version 1 | 87 | Not in stock | |||||
| BBa_K3590029 | Linker 1 version 2 | 87 | Not in stock | |||||
| BBa_K3590030 | Linker 1 version 3 | 87 | Not in stock | |||||
| BBa_K3590031 | Linker 1 version 4 | 87 | Not in stock | |||||
| BBa_K3590032 | Linker 2 version 1 | 105 | Not in stock | |||||
| BBa_K3590033 | Linker 2 version 2 | 105 | Not in stock | |||||
| BBa_K3590034 | Linker in front of a dockerin version 1 | 21 | Not in stock | |||||
| BBa_K3590035 | Linker in front of a dockerin version 2 | 21 | Not in stock | |||||
| BBa_K3831000 | BMC - pduA | 285 | ||||||
| BBa_K3831001 | BMC - pduB | 783 | ||||||
| BBa_K3831002 | BMC - pduJ | 276 | ||||||
| BBa_K3831003 | BMC - pduK | 696 | ||||||
| BBa_K3831004 | BMC - pduN | 270 | ||||||
| BBa_K3831013 | Ggggs linker | 15 | ||||||
| BBa_K3831016 | sfGFP | 714 | ||||||
| BBa_K3831017 | Localization tag for encapsulation in BMC | 24 | ||||||
| BBa_K3831019 | Degradation tag | 45 | ||||||
| BBa_K3831021 | mScarlet-I | 696 | ||||||
| BBa_K3831024 | cI repressor | 711 | ||||||
| BBa_K3831029 | Green fluorescent protein | 714 | ||||||
| BBa_K3831042 | GFP | 714 | ||||||
| BBa_K3831046 | PduN - reverse complement | 270 | ||||||
| BBa_K3831048 | PduK - reverse complement | 696 | ||||||
| BBa_K3831050 | PduJ - reverse complement | 276 | ||||||
| BBa_K3831052 | PduB - reverse complement | 783 | ||||||
| BBa_K3831054 | PduA - reverse complement | 285 | ||||||
| BBa_K4202000 | mazE(codon optimization for Bacillus subtilis) | 249 | ||||||
| BBa_K4202001 | mazF(codon optimization for Bacillus subtilis) | 421 | ||||||
| BBa_K4382000 | CglT | Beta-glucosidase | 1353 | |||||
| BBa_K4382001 | EG5C-1 | An endoglucanase, EG5C-1 | 996 | |||||
| BBa_K4382002 | DyP1B | Dye decolourising peroxidase (DyP1B) | 888 | |||||
| BBa_K4382003 | xynB | Beta-xylosidase (XynB) | 1602 | |||||
| BBa_K4382004 | Glucuronoxylan depolymerase - Xylanase C | 1269 | ||||||
| BBa_K4382005 | XynD | Arabinofuranoside - XynD | 1542 | |||||
| BBa_K4382006 | PelB-B2 | Endo Pectin Lyase - PelB-B2 | 1065 | Not in stock | ||||
| BBa_K4382007 | PelA | Pectin lyase - PelA | 1263 | |||||
| BBa_K4382008 | Pme | Pectin Methyl Esterase (Pme) | 1191 | |||||
| BBa_K4382010 | xynA | An endoxylanase - XynA | 642 | |||||
| BBa_K802006 | Surfactin generator for B. subtilis | 787 | Not in stock | |||||
| BBa_K802007 | Biofilm repressor for B. subtilis strains | 304 | It's complicated | |||||
| BBa_K802008 | Transcriptional Bacillus regulator lacI | 1193 | Not in stock | |||||
| BBa_K802009 | Sufactin generator and biofilm repressor for B. subtilis | 2300 | It's complicated | |||||
| BBa_K823020 | cat | chloramphenicol acetyltransferase (cat) | 645 | It's complicated | ||||
| BBa_K823031 | cotZ: B. subtilis spore crust protein (-2aa) | 435 | It's complicated | |||||
| BBa_K823032 | cotZ: B. subtilis spore crust protein | 441 | It's complicated | |||||
| BBa_K863121 | GFPmut3b | green fluorescent protein derived from jellyfish Aequeora victoria wild-type GFP (His-tag) | His6 | 750 | Not in stock |
DNA parts for building integration vector
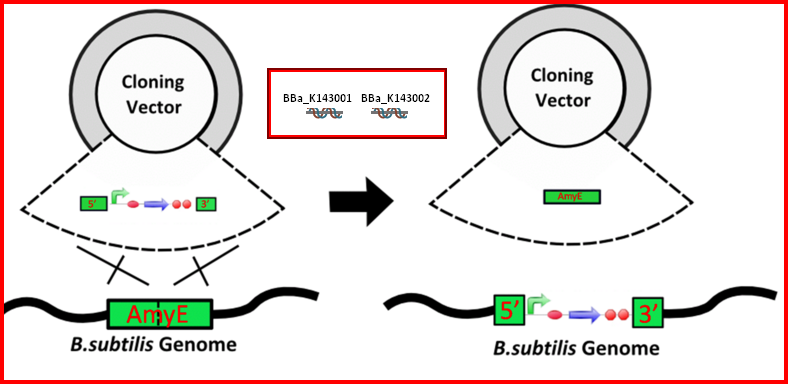
Any E. coli plasmid can be converted into an integration vector if one add two pieces of a a gene sequence that is homologous to a gene into B. subtilis genes, and an antibiotic resistance cassette or other screening method.
| The AmyE integration DNA parts (BBa_K143001 and BBa_K143002) are two parts that can be added to the 5' and 3' ends of a construct to allow integration into the B. subtilis genome. These parts have been successfully used within the parts BBa_K143079 and BBa_K143082 for integration. Integrated synthetic biological systems offer better genetic stability and more regulated copy number than plasmid-borne systems. For more information about these parts, please see [http://2008.igem.org/Team:Imperial_College 2008 Imperial College iGEM team wiki.] |
| Name | Description | Sequence | Length |
|---|---|---|---|
| BBa_K143001 | 5ĺ Integration Sequence for the amyE locus of B. subtilis | . . . aacacacaaattaaaaactggtctgatcga | 522 |
| BBa_K143002 | 3ĺ Integration Sequence for the amyE locus of B. subtilis | . . . tcgggcttaagcggttctcttccccattga | 1002 |
| BBa_K3831010 | Spacer_0 | . . . cttactctgttgaaaacgaatagataggtt | 40 |
| BBa_K3831011 | Spacer_1 | tgctcgtagtttacc | 15 |
| BBa_K3831012 | Spacer_5 | . . . agaatagtcaatcttcggaaatcccaggtg | 40 |
| BBa_K3831015 | Spacer_7 | taataaaaggtcccg | 15 |
| BBa_K3831023 | Spacer 03 | aaggaacggttattt | 15 |
| BBa_K3831026 | Spacer 02 | agattactactgata | 15 |
| BBa_K3831027 | Spacer 06 | ccgattctgagacgg | 15 |
| BBa_K3831028 | Spacer 04 | . . . aatacaggacccgaatcgtttcagttgcct | 40 |
| BBa_K3831038 | Spacer_SP1 | . . . ttaccacggatacagacagtgataatctta | 40 |
References
Given the number of available articles on B. subtilis, we only include some review articles here.
<biblio>
- Earl pmid=18467096
- Pavlendova pmid=18450217
- Sonenshein pmid=17982469
- Lopez pmid=17981078
- Aguilar pmid=17977783
- Irnov pmid=17381303
</biblio>