Protein domains
Notes on making protein domains: If you want to construct a protein doman, we recommend you use DNA synthesis . We recommend synthesis for the following reasons:
- Life is too short. DNA Synthesis is available to iGEM Teams. Check out the iGEM partner programs.
- You can have what you want. DNA Synthesis allows you to have exactly the sequence you want. You can optimize codons for your organism or manage mRNA stability with a tail.
- RFC #10 leaves an 8 bp scar site. To successfully make a protein, codons need to be in multiples of three (3) or a frameshift can be introduced. All RNA after a frameshift will be incorrectly translated, severely affecting or changing protein function.
- Scar sites can have charge. Strong charge will affect how your protein folds. The best way to eliminate issues with incorrect charge is to not have scar sites.
Please see our page on DNA Syntesis to find out more!
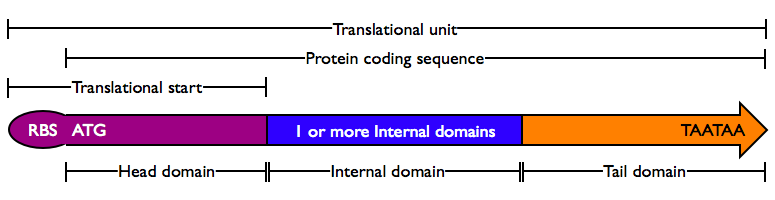
Protein domains encode portions of proteins and can be assembled together to form translational units, a genetic part spanning from translational initiation (the RBS) to translational termination (the stop codon).
There are several different types of protein domains.
- Head Domain: The Head domain consists of the start codon followed immediately by zero or more triplets specifiying an N-terminal tag, such as a protein export tag or lipoprotein binding tag. Head domains should begin with an
ATGstart codon and include codons 2 and 3 of the protein at a minimum. Examples of head domains includeATGstart codonATGstart codon and codons 2-3ATGstart codon and signal sequenceATGstart codon and affinity tag
- Internal Domains: Protein domains consist of a series of codon triplets coding for an amino acid sequence without a start codon or stop codon. Multiple Internal Domains can be fused. Examples of internal domains include
- DNA binding domains
- Dimerization domains
- Kinase domains
- Special Internal Domains: Short Domains with specific function may be separately categorized, but obey the same composition rules as normal internal domains. Examples of special internal domains include
- Linkers
- Cleavage sites
- Inteins
- Tail Domain: The C-terminus of a coding region consists of zero or more triplet codons, followed by a pair of TAA stop codons. In the simplest case, the stop codons terminate the protein with an Stop. More complex Tails may include degradation tags appropriate to the organism (i.e., with different degradation rates). Examples of Tail domain include
TAATAAstop codons- A degradation tag followed by
TAATAAstop codons - An affinity tag followed by
TAATAAstop codon
Unfortunately, the original BioBrick assembly standard, Assembly standard 10, does not support in-frame assembly of protein domains. (Assembly standard 10 creates an 8 bp scar between adjacent parts.) Therefore, it is recommended that you use an alternate approach to assemble protein domains together to make a translational unit. There are several possible approaches to assembling protein domains including direct synthesis (preferred because it creates no scars) as well as various assembly standards. Regardless of which standard you choose, we suggest that the resulting protein coding sequence or translational unit comply with the original BioBrick assembly standard so that your parts can be assembled with most of the parts in the Registry.
Protein coding sequences should be as follows
GAATTC GCGGCCGC T TCTAG [ATG ... TAA TAA] T ACTAGT A GCGGCCG CTGCAG
Note: Although most RBSs are currently specified as separate parts in the Registry, we are now moving to a new design in which the RBS and Head domain are combined into a single part termed a Translational start. The new design has the advantage of encapsulating both ribosome binding and translational initiation within a single part. Our working hypothesis is that the new design will reduce the likelihood of unexpected functional composition problems between the RBS and coding sequence.
| Design: Are you interested in designing a new protein domain part sequence? Here are some guidelines to help you design new protein domains. | |
| Help: A glossary, FAQ, and further reading on translational units, protein domains and protein coding sequences. |
Head domains
| Localization signals: Short protein sequences that "tell" a cell where to send a protein. For example, a localization sequence might send a protein to the nucleus or mark it for secretion outside of the cell. |
Internal Domains
| DNA binding domains: Domains that bind to specific DNA sequences, called operators. | |
| Reporter protein domains: Reporters are protein domains or protein coding sequences that can be used to measure gene expression or other intracellular events. Reporters generally produce a measurable signal such as fluorescence, color, or luminescence. | |
| Miscellaneous protein domains: Other internal protein domains. |
Special Internal Domains
| Linkers: Short protein sequences used to link or fuse two protein domains together. | |
| Cleavage sites: Short protein sequences or motifs that are recognized by site-specific proteases that then cut a particular peptide bond. |
Tail domains
| Degradation tags: Short protein sequences or protein domains that target a protein for degradation by cellular machinery thereby increasing protein turnover and decreasing protein half-life. Since most degradation tags rely on endogenous cellular machinery for degradation, degradation tags are specific to particular chassis. |
Head or tail domains
| Affinity tags: Short protein sequences or protein domains that can bind to antibody, metal ion, or other substrate. Generally, affinity tags are fused to proteins to enable purification via binding to an immobilized ligand or quantification via western blot. |