Part:BBa_K2560123
Phytobrick version of pTet
This is the Phytobrick version of the promoter pTet and was build as a part of the Marburg Collection. Instructions of how to use the Marburg Collection are provided at the bottom of the page.
Promoters
Promoters are genetic modules were the RNA polymerase is recruited to start RNA transcription. They are divided in two groups: constitutive promoters which transcribe RNA permanently and inducible promoters which start the transcription as a response to a stimulus.
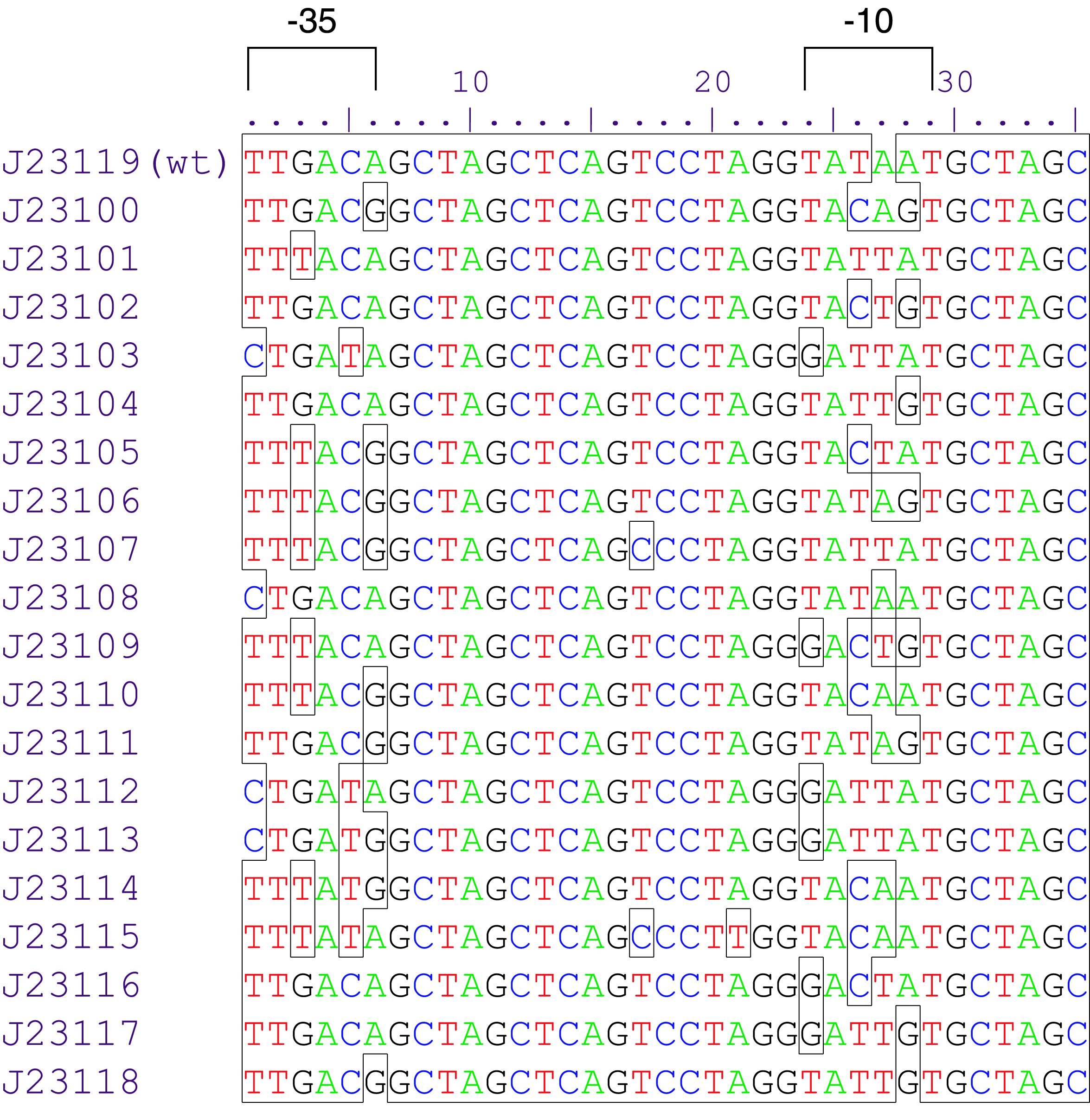
Inducible promoters can be regulated by transcription activation or repression. To start the RNA transcription the RNA polymerase complex is not sufficient. Therefore sigma70 factors are required. The sigma70 factor binds to the Pribnow box – two motifs -10 and -35bp upstream the CDS – recruiting the RNA transcription complex enabling the transcription. Based on this knowledge a collection of synthetic constitutive promoters have been developed by Chris Anderson and made available on the iGEM repository.
Characterization
Anderson Promoters
Variant Lux (au) K2560131 (Dummy) 0.025 K2560019 (J23103) 0.032 K2560026 (J23113) 0,038 K2560023 (J23109) 0,052 K2560009 (J23104) 0,058 K2560029 (J23117) 0,090 K2560025 (J23111) 0,098 K2560028 (J23116) 0,134 K2560021 (J23107) 0,136 K2560027 (J23114) 0,163 K2560024 (J23110) 0,169 K2560018 (J23102) 0,245 K2560030 (J23118) 0,348 K2560020 (J23105) 0,387 K2560015 (J23115) 0,398 K2560014 (J23106) 0,502 K2560017 (J23101) 0,510 K2560022 (J23108) 0,768 K2560007 (J23100) 1
We started by measuring the promoter strength of the Anderson Promoter library in V. natriegens. Firstly, we assembled 19 test plasmids with golden-gate-assembly and measured their expression strength, following our selfmade workflows. The results are shown in Figure 1. We observed an even distribution of the tested promoters throughout the dynamic range. The strongest promoter K2560007 (J23100) yielded 40 fold stronger signal than the promoter dummy and was used as a reference to calculate relative promoter strengths.
The test constructs were built with dummy connectors which did not possess insulator elements. We assume that this resulted in additional expression caused by transcription throughout the rest of the plasmid, e.g. ori and antibiotic resistance. This is thought to add the same extent of signal to all measured promoters thus reducing the overall dynamic range. To further evaluate this assumption, we could repeat this experiment with one of our insulators instead of the dummy connector.
Inducible Promoters
In addition to constitutive promoters, the Marburg Collection contains two inducible promoters, pTet and pTrc. For all experiments with inducible promoters, we added the respective inducer concentration to the preculture as well as to the main culture to ensure constant expression.The first experiments were performed with the pTet promoter that can be induced by the tetracycline derivative anhydrotetracycline (ATc). ATc is much less cytotoxic but still capable of binding and altering the structure of the repressor TetR, leading to release of the promoter and enabling transcription. To measure the dose response behavior of the pTet, we made a dilution series of ATc. Following the recommendation of our advisors (Stefano Vecchione), we started with the concentration commonly used in E. coli, started with the concentration (100 ng/mL). The starting concentration was diluted twofold in 20 subsequent steps. Our results are shown in figure xxx. The absence of bars for the four highest concentrations means that the cultures did not reach an OD of 0.2 in the seven hours of the measurement. Remarkably, we observed reasonable growth of those same cultures in the preculture already induced with the identical amount of ATc. Knowing that luminescence is produced at the end of an enzymatic cascade, starting with intermediates of the phospholipid metabolism ( Meighen 1991), we reckon that very strong induction could decrease the fitness of cells and that after dilution in room temperature medium, strained cells are not able to recover from the stationary phase. However, we only observed this phenomenon in experiments with pTet, although we obtained higher signals for the strongest constitutive promoters as well as for the highly induced pTrc. We checked for toxicity of ATc but could not see a measurable effect (figurexxxxx). Another possibility is that TetR interacts with components inside the cell and that high ATc increases these interactions. Blast searches of TetR against the genome of V. natriegens identified one protein that shares some homology with the N-terminal part of TetR which could result in cross talk between the host and the inducible promoter.
All measured data were normalized to the strongest constitutive promoter J23100. Saturation occurred at a dilution of 2^6 (~ 1.6 ng/mL) and an exponential reduction of luminescence signal can be observed for higher dilutions. In the absence of ATc, the signal is twelve fold lower compared to saturation.
pTet allows relatively tight control of gene expression and is therefore well suited for driving the expression of potentially toxic proteins. On the other hand, we were not able to induce strong expression that can compete with strong constitutive promoters or the fully induced pTrc.
pTrc is the second tested inducible promoter. It contains lac operator sites and is therefore regulated by the repressor LacI which is constitutively expressed from a downstream gene. pTrc can be induced Isoopropyl-β-D-thiogalactopyranosid (IPTG), a chemical derivative of lactose ( Camsund et al.2014). Similar to our experiments with pTet, we made a dilution series starting with the commonly used IPTG concentration for E. coli 0.5 mM. We observed a five fold induction and a saturation that occurred at a dilution of 2^5 (~15 µM). The strongest expression is similar to the expression gained from the strongest constitutive promoter J23100 while the expression in the absence of inducer equals medium strong promoters. As a consequence, we do not recommend using pTrc in constructs where a tight control of gene expression is desired. However, pTrc is well suited when strong expression is required.
Taking the results of both inducible promoters into account, we made two observation. In both cases, the dynamic range is smaller compared to E. coli and the inducer concentration that facilitates saturation is 32 and 64 fold lower for pTrc and pTet, respectively, than the concentration that is typically used for E. coli. A possible explanation could be found in the fast growth of V. natriegens which might result in a lower concentration of the repressor proteins in the cells, finally leading to a less restricted control of the negatively regulated promoters. However, we do not have experimental support for our idea.
Usage and Biology
Marburg 2018 characterized this part in Vibrio natriegens using the lux operon of Photorhabdus luminescens (BBa_K2560051). The parts sequence was verified by Sanger sequencing.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Marburg Toolbox
We proudly present the Marburg Collection, a novel golden-gate-based toolbox containing various parts that are compatible with the PhytoBrick system and MoClo. Compared to other bacterial toolboxes, the Marburg Collection shines with superior flexibility. We overcame the rigid paradigm of plasmid construction - thinking in fixed backbone and insert categories - by achieving complete de novo assembly of plasmids.
36 connectors facilitate flexible cloning of multigene constructs and even allow for the inversion of individual transcription units. Additionally, our connectors function as insulators to avoid undesired crosstalk.
The Marburg Collection contains 123 parts in total, including:
inducible promoters, reporters, fluorescence and epitope tags, oris, resistance cassettes and genome engineering tools. To increase the value of the Marburg Collection, we additionally provide detailed experimental characterization for V. natriegens and a supportive software. We aspire availability of our toolbox for future iGEM teams to empower accelerated progression in their ambitious projects.
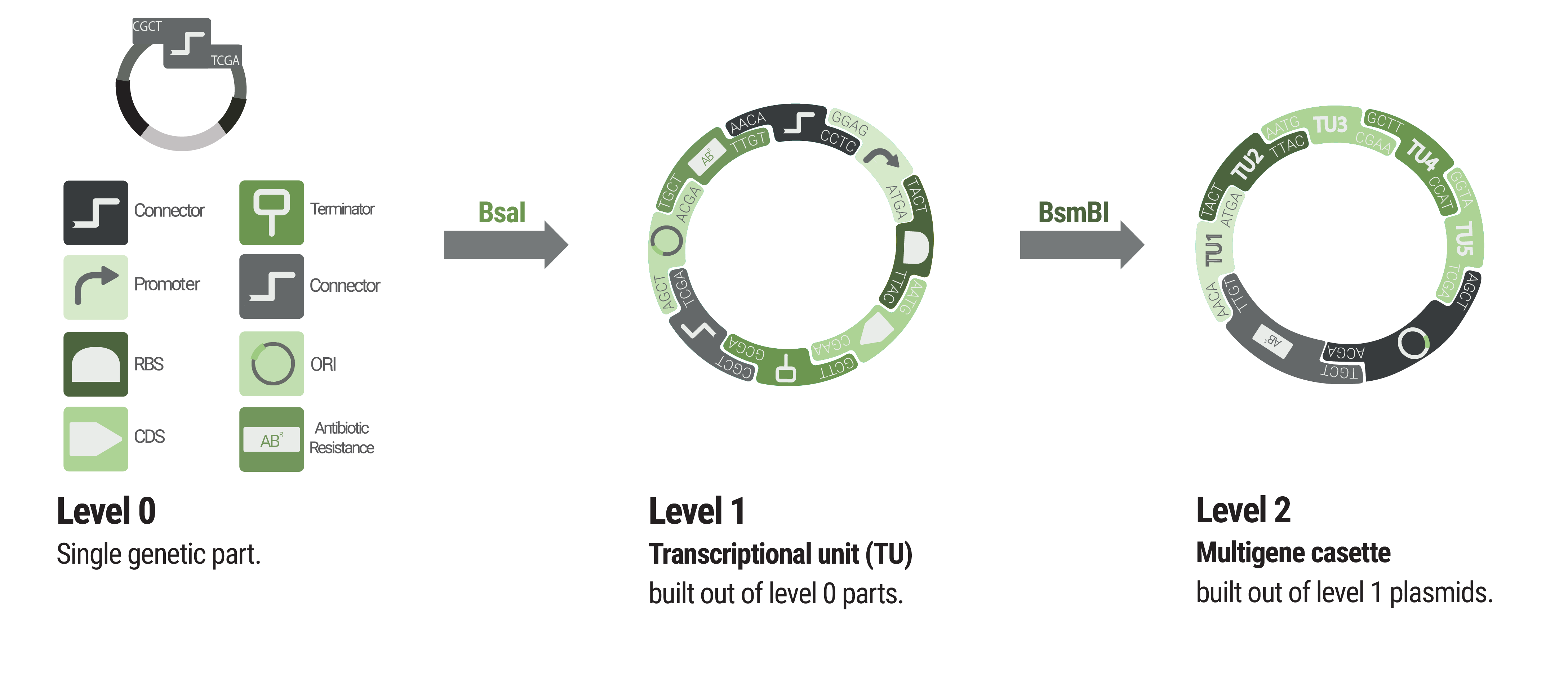
Basic building blocks like promoters or terminators are stored in level 0 plasmids. Parts from each category of our collection can be chosen to built level 1 plasmids harboring a single transcription unit. Up to five transcription units can be assembled into a level 2 plasmid.
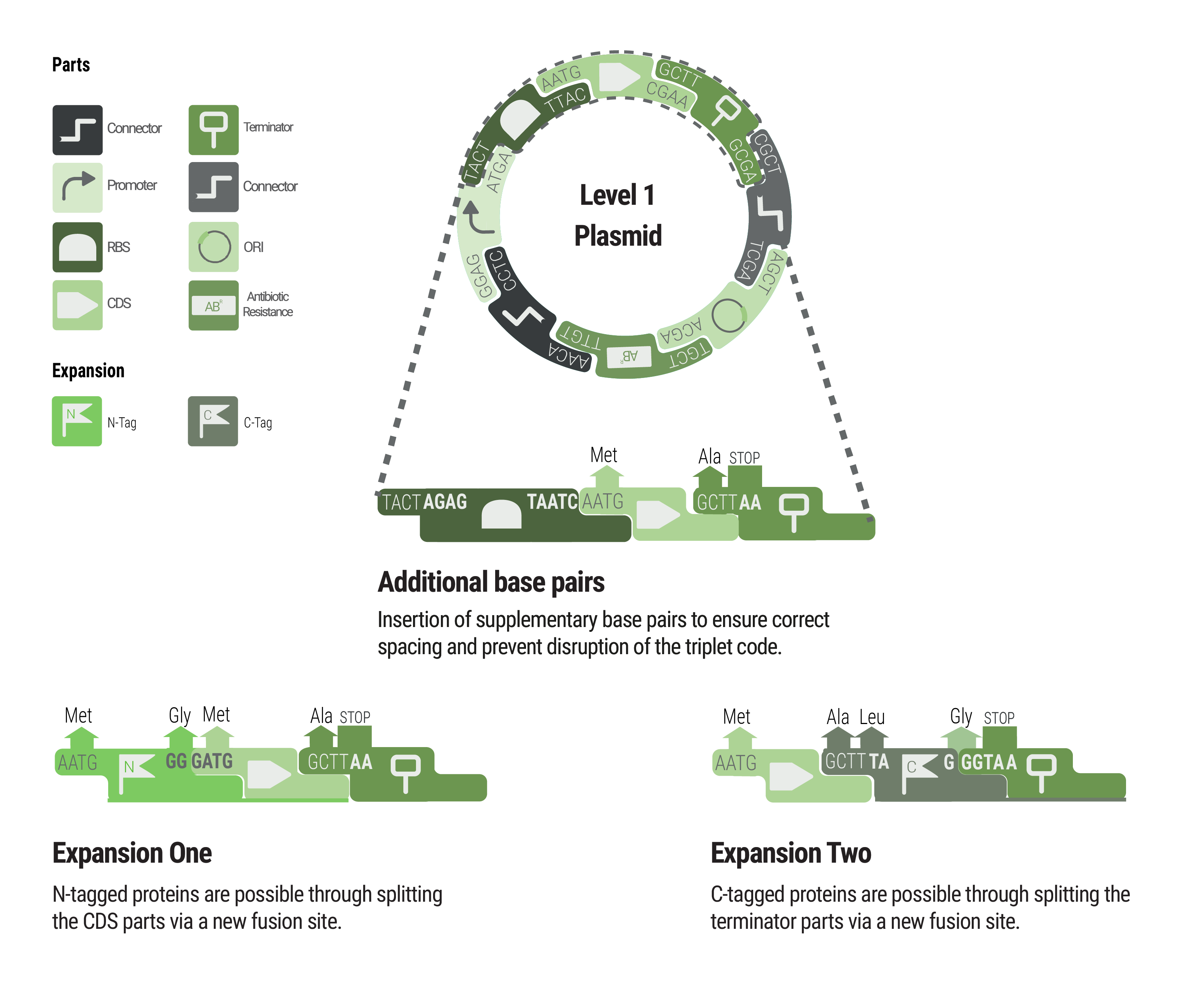
Between some parts, additional base pairs were integrated to ensure correct spacing and to maintain the triplet code. We expanded our toolbox by providing N- and C- terminal tags by creating novel fusions and splitting the CDS and terminator part, respectively.
Parts of the Marburg Toolbox

- K2560011 (5'Connector Dummy)
- K2560055
(1-6
Connector) - K2560065 (5'Con1)
- K2560066 (5'Con2)
- K2560067 (5'Con3)
- K2560068 (5'Con4)
- K2560069 (5'Con5)
- K2560075 (5'Con1
Short Res) - K2560076 (5'Con2
Short) - K2560077 (5'Con3
Short) - K2560078 (5'Con4
Short) - K2560079 (5'Con5
Short) - K2560095 (5'Con1 inv)
- K2560096 (5'Con2 inv)
- K2560097 (5'Con3 inv)
- K2560098 (5'Con4 inv)
- K2560099 (5'Con5 inv)
- K2560105 (5'Con5 inv
Ori) - K2560107 (5'Con1
Res)

- K2560007 (J23100)
- K2560009 (J23104)
- K2560014 (J23106)
- K2560015 (J23115)
- K2560017 (J23101)
- K2560018 (J23102)
- K2560019 (J23103)
- K2560020 (J23105)
- K2560021 (J23107)
- K2560022 (J23108)
- K2560023 (J23109)
- K2560024 (J23110)
- K2560025 (J23111)
- K2560026 (J23113)
- K2560027 (J23114)
- K2560028 (J23116)
- K2560029 (J23117)
- K2560030 (J23118)
- K2560031 (J23119)
- K2560123
(pTet) - K2560124 (pTrc)
- K2560131 (Promoter Dummy)

- K2560012 (3'Connector Dummy)
- K2560070 (3'Con1)
- K2560071 (3'Con2)
- K2560072 (3'Con3)
- K2560073 (3'Con4)
- K2560080 (3'Con5 Ori)
- K2560100 (3'Con1 inv
Short) - K2560101 (3'Con2 inv
Short) - K2560102 (3'Con3 inv
Short) - K2560103 (3'Con4 inv
Short) - K2560104 (3'Con5 inv
Short) - K2560106 (3'Con1 inv
Short Res) - K2560108 (3'Con1 inv)
- K2560109 (3'Con1 inv
Res) - K2560110 (3'Con2 inv)
- K2560111 (3'Con3 inv)
- K2560112 (3'Con4 inv)
- K2560113 (3'Con5 inv)

- K2560048 (Cam. Res. RFP)
- K2560056
(Kan. Res. (pSB3K3) RFP) - K2560057
(Kan. Res. (pSB3K3) GFP) - K2560058
(Tet. Res. (pSB3T5) RFP) - K2560059
(Tet. Res. (pSB3T5) GFP) - K2560125 (Carb. Res. RFP)
- K2560126 (Carb. Res. GFP)
- K2560127 (Carb. Res. into BBa_K2560002)
- K2560132 (Cam. Res. into BBa_K2560002)
- K2560133
(Kan. Res. into BBa_K2560002) - K2560134
(Tet. Res. into BBa_K2560002)
Tags and Entry Vectors