Part:BBa_K2560034
Phytobrick version of BBa_B0010
This is the Phytobrick version of the Terminator BBa_B0010 and was build as a part of the Marburg Collection. Instructions of how to use the Marburg Collection are provided at the bottom of the page.
Overview
Terminators are sequences downstream CDSs promoting dissociation of the transcriptional complex. In prokaryotes two kind of transcription terminators are reported. The first ones are Rho-independent terminators. This sequences build up hairpin structures during transcription disrupting the transcriptional complex. The mechanism of disruption is hypothesized to occur through allosteric effects of the hairpin binding and competitive kinetics ( Farnham PJ and Platt T.,1981) The other group are Rho-dependent terminators which require Rho and ATP. This terminators are found in bacteria and phages. The Rho factor binds to a cytosine-rich sequence on the mRNA where it hydrolyzes ATP, translocates down the mRNA and stimulates the dissociation of the transcription complex when contact is established ( Banerjee et al.2006).
Characterization
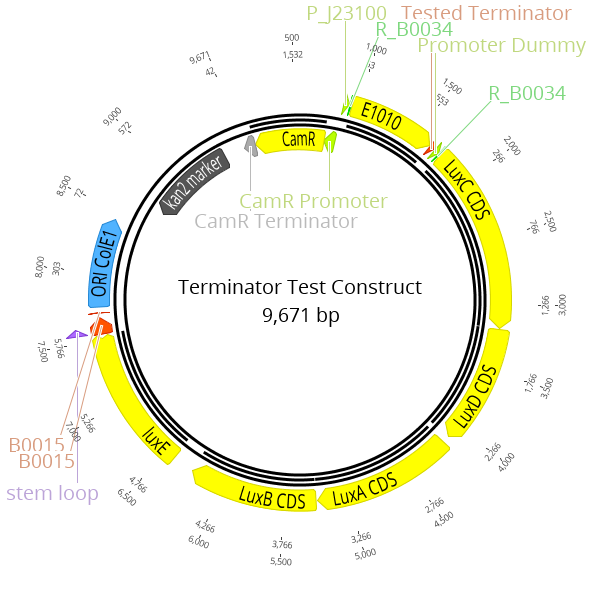
LVL2 plasmids were created for these experiments consisting of a RFP transcription unit with the strong constitutive promoter J23100, followed by the lux operon with the promoter dummy. The terminator located at the 3' end of the RFP transcription unit is the part which is characterized in this experiment.
The Marburg Collection contains five terminators plus one terminator dummy that can be used as a placeholder. To obtain experimental data for this category of parts, we built a set of terminator test constructs to measure the extent of transcriptional readthrough and therefore the strength of a terminator. The terminator test constructs are built as LVL2 plasmid with our toolbox. The strongest constitutive promoter K2560007 (J23100) drives the expression of RFP which is the first transcription unit. The Lux operon is placed downstream with the promoter dummy instead of an active promoter. Both transcription units are separated by the terminator, which is the focus of this characterization, downstream of the RFP CDS.
With this setup, transcriptional activity of the RFP reporter is blocked by the terminator. Therefore the measured luminescence signal can be seen as an indicator for the efficiency of the terminator.
As discussed previously, RFP is not suitable for precise quantitative characterizations. Therefore we did not calculate ratios of the reporter upstream and downstream of the tested reporter as was described in previous experiments ( Chen et al.2013). However, we used the existence of an RFP signal as an control for the correctly assembled test constructs and for the activity of the promoter driving RFP.
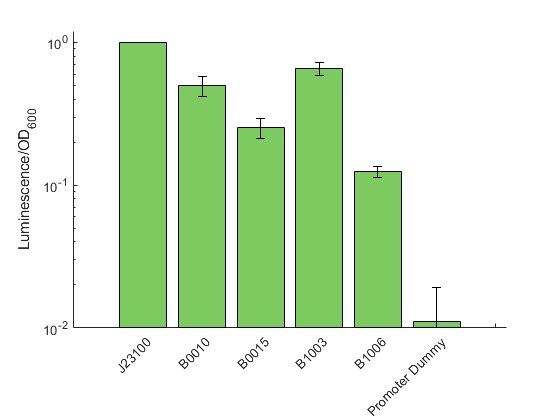
The data shown in figure 2 were acquired and analyzed following our novel workflow described on the measurement page . Like in all previous experiments, the obtained raw data for each sample were normalized over the construct J23100 from the promoter characterization. The strongest signal was observed for B1002 and B0010 with a relative signal 0.65 and 0.50 respectively, suggesting these two terminators as rather inefficient. In contrast, we could show a signal reduction four and eight fold for B0015 and B1006, respectively.
By comparing our data to the characterization provided in the iGEM registry for E. coli, we can show that the general trend is similar for both organisms. Exemplary, the terminator with the highest described efficiency in E. coli (B1006) also was found to reduce the luminescence signal most in our experiments.
In general, we found stronger signals for the reporter downstream of the terminator than what was described for E. coli. However, it has to be noted, that we used the highly sensitive reporter Lux instead of a fluorescent protein. Therefore we assume that we were able to detect a higher degree of transcriptional readthrough, which would not be distinguishable from the background when using a fluorescence reporter.
However, we cannot exclude species specific differences that cause a generally higher degree of transcriptional readthrough over terminators for V. natriegens compared to E. coli
In addition to the terminators shown in figure 2 we also measured a test construct possessing B1003. The, by far, lowest signal was found for B1003 with a 15000 fold reduction of the signal. To ensure that is extremely weak signal is authentic and not caused by cross talk from neighbouring wells, we tested this sample in an otherwise empty 96 well plate and confirmed the general existence of a signal. However, we can not exclude the presence of single mutations within the lux operon dramatically diminishing the overall generation of luminescence. This result was not displayed in figure 2 to omit extreme stretching of the Y-Axis thus loosing visual information for the other tested terminators.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Marburg Toolbox
We proudly present the Marburg Collection, a novel golden-gate-based toolbox containing various parts that are compatible with the PhytoBrick system and MoClo. Compared to other bacterial toolboxes, the Marburg Collection shines with superior flexibility. We overcame the rigid paradigm of plasmid construction - thinking in fixed backbone and insert categories - by achieving complete de novo assembly of plasmids.
36 connectors facilitate flexible cloning of multigene constructs and even allow for the inversion of individual transcription units. Additionally, our connectors function as insulators to avoid undesired crosstalk.
The Marburg Collection contains 123 parts in total, including:
inducible promoters, reporters, fluorescence and epitope tags, oris, resistance cassettes and genome engineering tools. To increase the value of the Marburg Collection, we additionally provide detailed experimental characterization for V. natriegens and a supportive software. We aspire availability of our toolbox for future iGEM teams to empower accelerated progression in their ambitious projects.
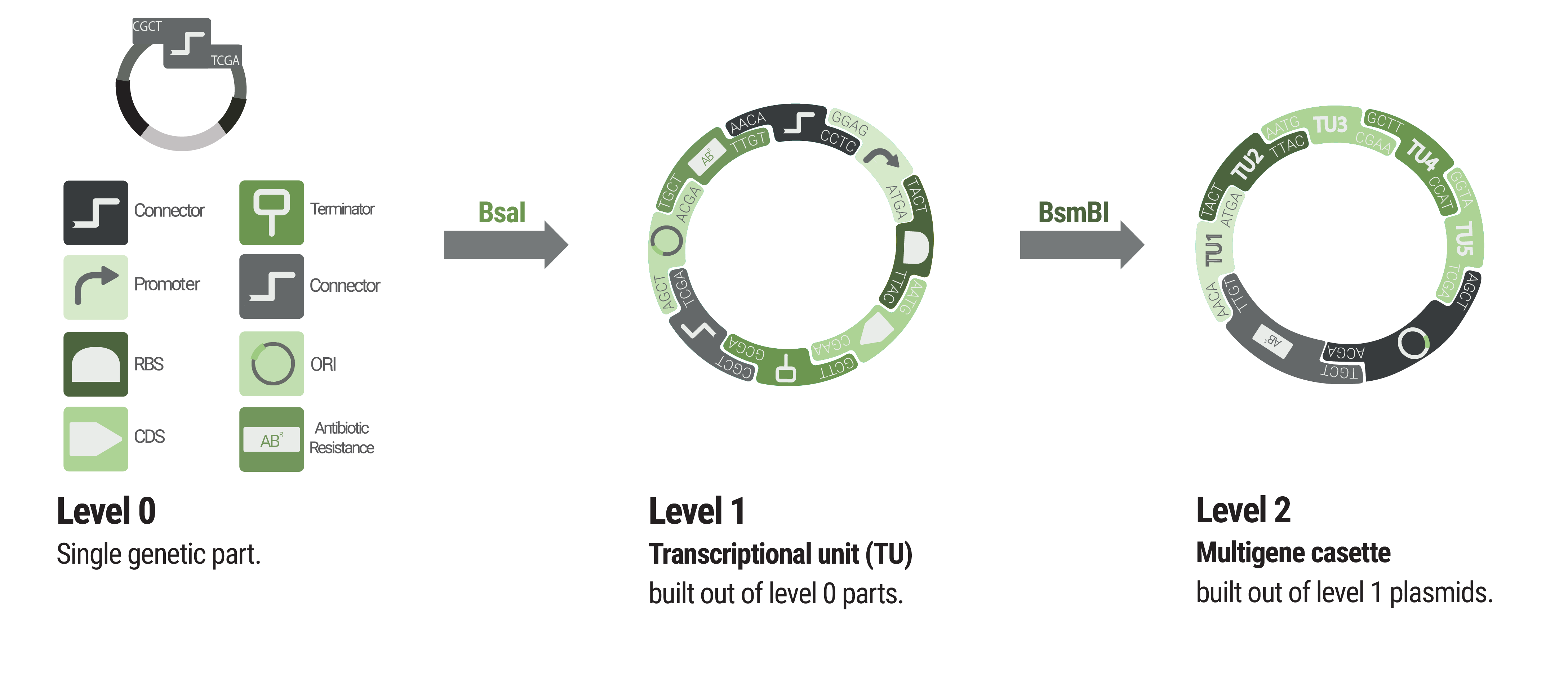
Basic building blocks like promoters or terminators are stored in level 0 plasmids. Parts from each category of our collection can be chosen to built level 1 plasmids harboring a single transcription unit. Up to five transcription units can be assembled into a level 2 plasmid.
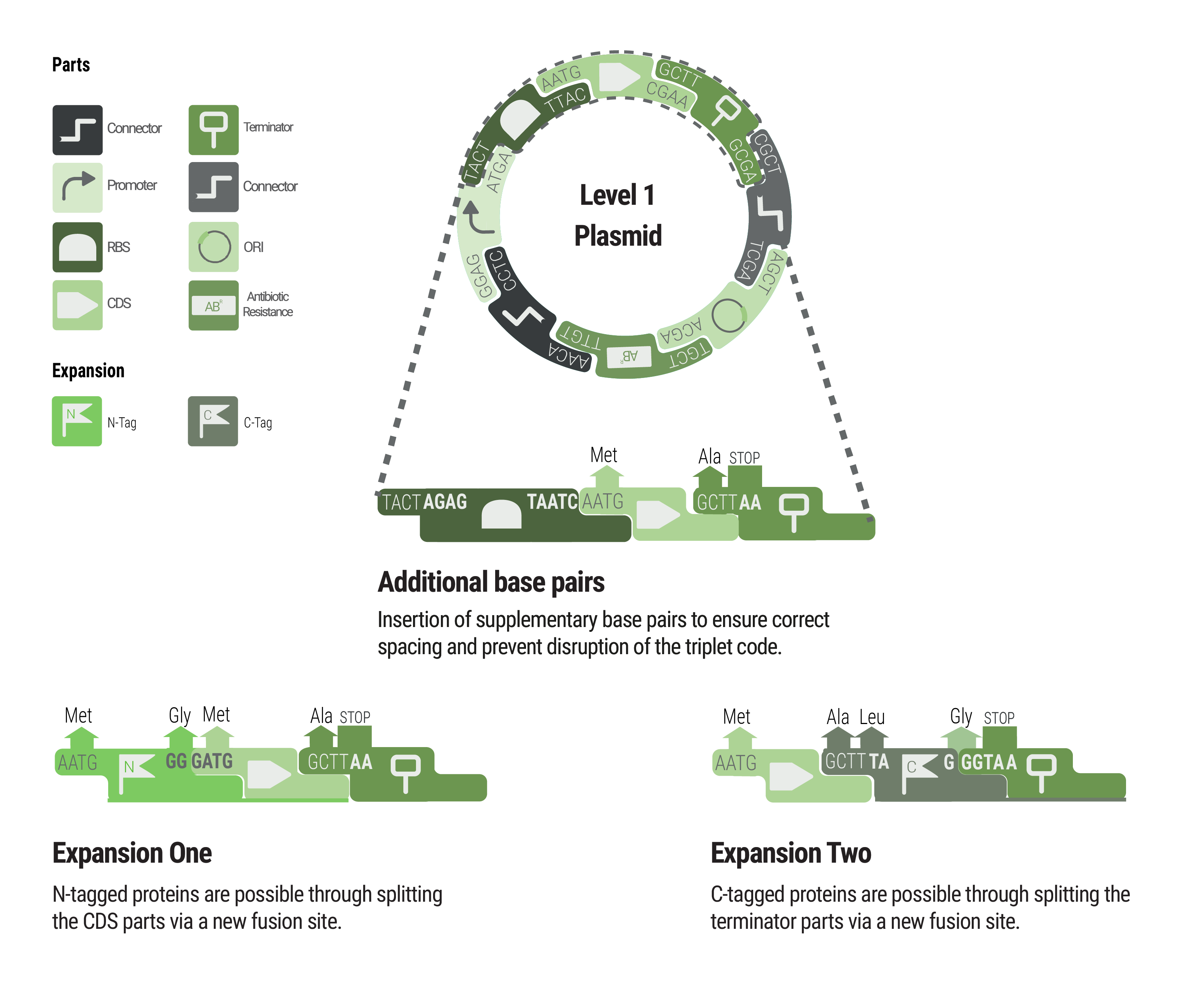
Between some parts, additional base pairs were integrated to ensure correct spacing and to maintain the triplet code. We expanded our toolbox by providing N- and C- terminal tags by creating novel fusions and splitting the CDS and terminator part, respectively.
Parts of the Marburg Toolbox

- K2560011 (5'Connector Dummy)
- K2560055
(1-6
Connector) - K2560065 (5'Con1)
- K2560066 (5'Con2)
- K2560067 (5'Con3)
- K2560068 (5'Con4)
- K2560069 (5'Con5)
- K2560075 (5'Con1
Short Res) - K2560076 (5'Con2
Short) - K2560077 (5'Con3
Short) - K2560078 (5'Con4
Short) - K2560079 (5'Con5
Short) - K2560095 (5'Con1 inv)
- K2560096 (5'Con2 inv)
- K2560097 (5'Con3 inv)
- K2560098 (5'Con4 inv)
- K2560099 (5'Con5 inv)
- K2560105 (5'Con5 inv
Ori) - K2560107 (5'Con1
Res)

- K2560007 (J23100)
- K2560009 (J23104)
- K2560014 (J23106)
- K2560015 (J23115)
- K2560017 (J23101)
- K2560018 (J23102)
- K2560019 (J23103)
- K2560020 (J23105)
- K2560021 (J23107)
- K2560022 (J23108)
- K2560023 (J23109)
- K2560024 (J23110)
- K2560025 (J23111)
- K2560026 (J23113)
- K2560027 (J23114)
- K2560028 (J23116)
- K2560029 (J23117)
- K2560030 (J23118)
- K2560031 (J23119)
- K2560123
(pTet) - K2560124 (pTrc)
- K2560131 (Promoter Dummy)

- K2560012 (3'Connector Dummy)
- K2560070 (3'Con1)
- K2560071 (3'Con2)
- K2560072 (3'Con3)
- K2560073 (3'Con4)
- K2560080 (3'Con5 Ori)
- K2560100 (3'Con1 inv
Short) - K2560101 (3'Con2 inv
Short) - K2560102 (3'Con3 inv
Short) - K2560103 (3'Con4 inv
Short) - K2560104 (3'Con5 inv
Short) - K2560106 (3'Con1 inv
Short Res) - K2560108 (3'Con1 inv)
- K2560109 (3'Con1 inv
Res) - K2560110 (3'Con2 inv)
- K2560111 (3'Con3 inv)
- K2560112 (3'Con4 inv)
- K2560113 (3'Con5 inv)

- K2560048 (Cam. Res. RFP)
- K2560056
(Kan. Res. (pSB3K3) RFP) - K2560057
(Kan. Res. (pSB3K3) GFP) - K2560058
(Tet. Res. (pSB3T5) RFP) - K2560059
(Tet. Res. (pSB3T5) GFP) - K2560125 (Carb. Res. RFP)
- K2560126 (Carb. Res. GFP)
- K2560127 (Carb. Res. into BBa_K2560002)
- K2560132 (Cam. Res. into BBa_K2560002)
- K2560133
(Kan. Res. into BBa_K2560002) - K2560134
(Tet. Res. into BBa_K2560002)
Tags and Entry Vectors
| None |