Part:BBa_K2560118
Phytobrick version of 5a BBa_M0050 SsrA degradation tag
This is the Phytobrick version of the tag M0050 SsrA degradation tag and was build as a part of the Marburg Collection. Instructions of how to use the Marburg Collection are provided at the bottom of the page.
Overview
Protein tags are peptides genetically fused to the proteins of interest. There are many different kinds of tags developed for for different purposes. Protein purification tags are peptide chains with a high affinity to a certain matrix like the histidine affinity tag binding to a Ni2+ matrix. This and other purification tags are used to separate the tagged protein from cell debris ( Kimple et al.2013). Another kind of tags that can be used in a similar way are epitope tags. Here a certain epitope is fused to the protein to be detected by the corresponding antibody. This technique can be used to purify the proteins or to label them in vivo with fluorescent labeled antibodies ( Brizzard B.,2008). Nevertheless for labeling experiments normally the proteins of interest are tagged directly by fluorescent tags. This method can be used to study protein localization, interactions and dynamics ( Crivat G, Taraska JW. ,2012). Degradation tags are another group of tags used to label the proteins in a well recognizable way for proteases reducing their half-life significantly. This makes this tags interesting for synthetic biology enabling the experimenter to influence protein abundances, degradation rates and consequently the cell’s respond ( Cameron DE, Collins J. ,2014).
Characterization
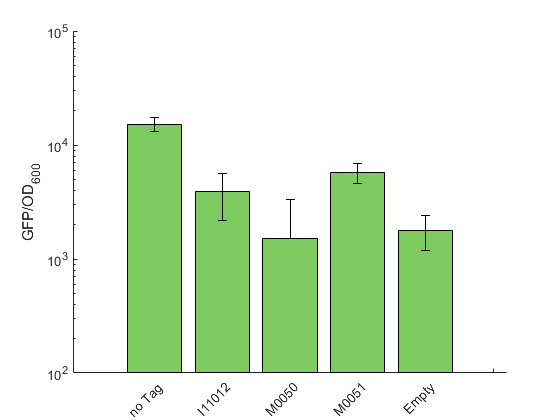
Our toolbox contains the three degradation tags K2560118 (M0050 5a), K2560119 (M0051 5a) and K2560117 (I11012 5a). Similar to our RBS experiments, we could not use the lux operon as a reporter because the degradation tag is only added to the C-terminal end of the last enzyme encoded in the operon. Therefore, we again used sfGFP as reporter and test constructs were designed with one of the tags fused to the CDS of sfGFP. A non-tagged sfGFP construct serves as the reference.
As expected, appending a degradation tag to a protein decreases its concentration. The strongest decrease and therefore the highest degradation was shown for K2560118 (M0050 5a) with a signal that is not distinguishable from the non-sfGFP expressing control. The second strongest signal reduction was shown for K2560117 (I11012 5a), followed by K2560119 (M0051 5a), which showed the least efficient degradation (figure 1). Our results are in qualitative accordance with the description of these tags for E. coli, which are provided in the registry.
The tested degradation tags belong to the family of SsrA degradation tags. Naturally, they help to degrade incompletely translated proteins by labeling them for Clp proteases (Farrell et al. 2015). We checked for the presence of ClpP, one of the proteases involved in degrading SsrA tagged proteins in E. coli and found a highly homologous protein in V. natriegens. Therefore we assume that the mechanism of Clp mediated degradation is conserved between both organisms, which is in accordance with our results.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Marburg Toolbox
We proudly present the Marburg Collection, a novel golden-gate-based toolbox containing various parts that are compatible with the PhytoBrick system and MoClo. Compared to other bacterial toolboxes, the Marburg Collection shines with superior flexibility. We overcame the rigid paradigm of plasmid construction - thinking in fixed backbone and insert categories - by achieving complete de novo assembly of plasmids.
36 connectors facilitate flexible cloning of multigene constructs and even allow for the inversion of individual transcription units. Additionally, our connectors function as insulators to avoid undesired crosstalk.
The Marburg Collection contains 123 parts in total, including:
inducible promoters, reporters, fluorescence and epitope tags, oris, resistance cassettes and genome engineering tools. To increase the value of the Marburg Collection, we additionally provide detailed experimental characterization for V. natriegens and a supportive software. We aspire availability of our toolbox for future iGEM teams to empower accelerated progression in their ambitious projects.
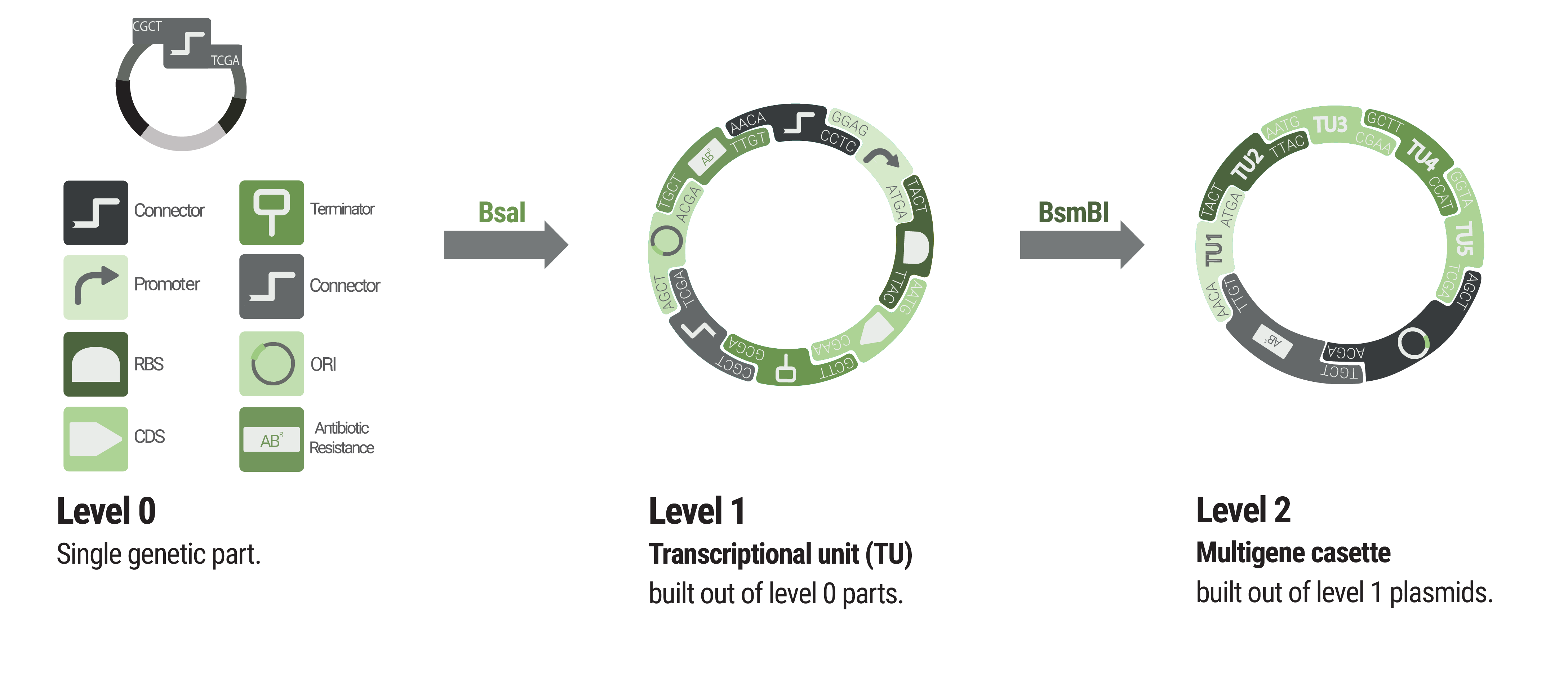
Basic building blocks like promoters or terminators are stored in level 0 plasmids. Parts from each category of our collection can be chosen to built level 1 plasmids harboring a single transcription unit. Up to five transcription units can be assembled into a level 2 plasmid.
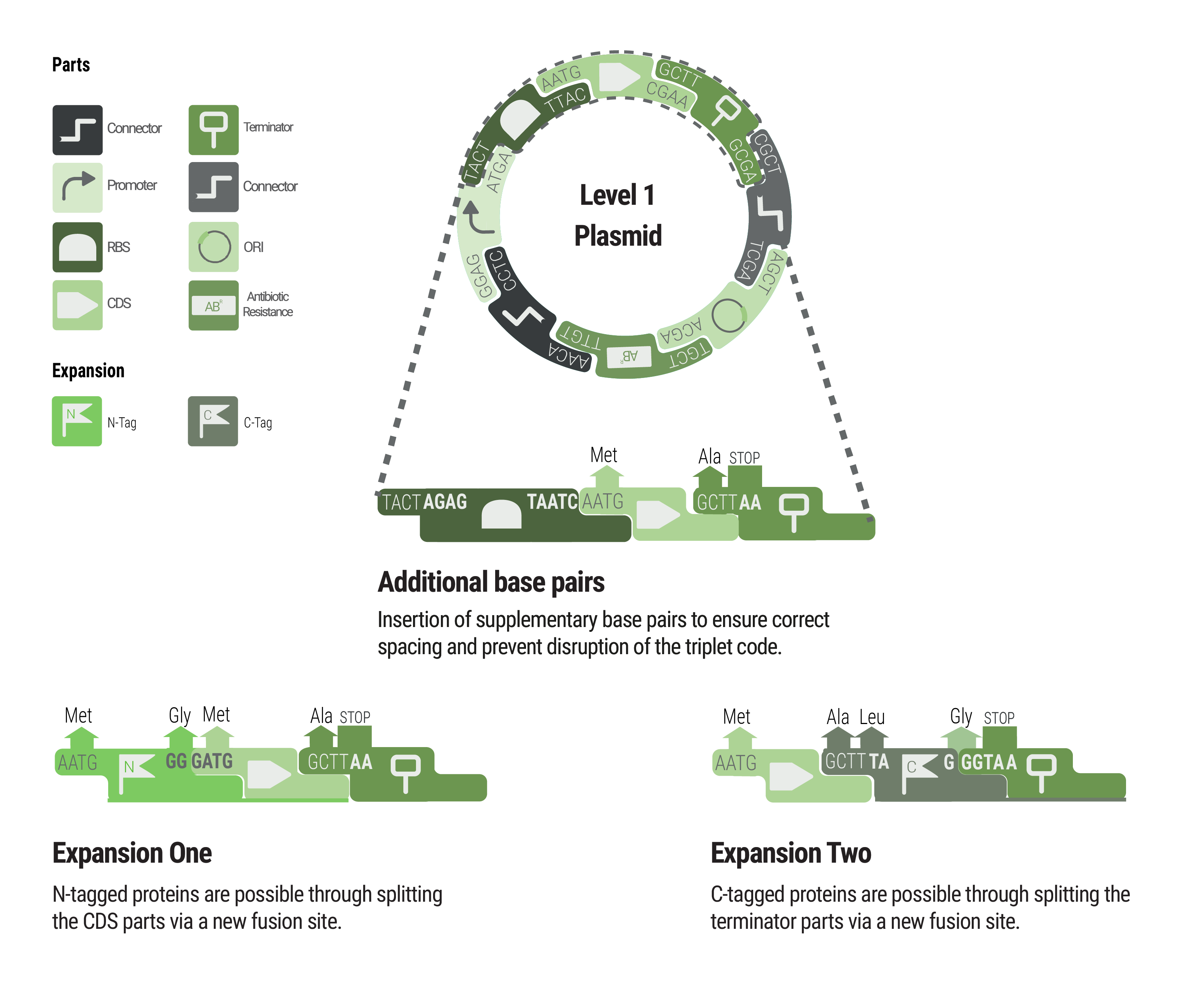
Between some parts, additional base pairs were integrated to ensure correct spacing and to maintain the triplet code. We expanded our toolbox by providing N- and C- terminal tags by creating novel fusions and splitting the CDS and terminator part, respectively.
Parts of the Marburg Toolbox

- K2560011 (5'Connector Dummy)
- K2560055
(1-6
Connector) - K2560065 (5'Con1)
- K2560066 (5'Con2)
- K2560067 (5'Con3)
- K2560068 (5'Con4)
- K2560069 (5'Con5)
- K2560075 (5'Con1
Short Res) - K2560076 (5'Con2
Short) - K2560077 (5'Con3
Short) - K2560078 (5'Con4
Short) - K2560079 (5'Con5
Short) - K2560095 (5'Con1 inv)
- K2560096 (5'Con2 inv)
- K2560097 (5'Con3 inv)
- K2560098 (5'Con4 inv)
- K2560099 (5'Con5 inv)
- K2560105 (5'Con5 inv
Ori) - K2560107 (5'Con1
Res)

- K2560007 (J23100)
- K2560009 (J23104)
- K2560014 (J23106)
- K2560015 (J23115)
- K2560017 (J23101)
- K2560018 (J23102)
- K2560019 (J23103)
- K2560020 (J23105)
- K2560021 (J23107)
- K2560022 (J23108)
- K2560023 (J23109)
- K2560024 (J23110)
- K2560025 (J23111)
- K2560026 (J23113)
- K2560027 (J23114)
- K2560028 (J23116)
- K2560029 (J23117)
- K2560030 (J23118)
- K2560031 (J23119)
- K2560123
(pTet) - K2560124 (pTrc)
- K2560131 (Promoter Dummy)

- K2560012 (3'Connector Dummy)
- K2560070 (3'Con1)
- K2560071 (3'Con2)
- K2560072 (3'Con3)
- K2560073 (3'Con4)
- K2560080 (3'Con5 Ori)
- K2560100 (3'Con1 inv
Short) - K2560101 (3'Con2 inv
Short) - K2560102 (3'Con3 inv
Short) - K2560103 (3'Con4 inv
Short) - K2560104 (3'Con5 inv
Short) - K2560106 (3'Con1 inv
Short Res) - K2560108 (3'Con1 inv)
- K2560109 (3'Con1 inv
Res) - K2560110 (3'Con2 inv)
- K2560111 (3'Con3 inv)
- K2560112 (3'Con4 inv)
- K2560113 (3'Con5 inv)

- K2560048 (Cam. Res. RFP)
- K2560056
(Kan. Res. (pSB3K3) RFP) - K2560057
(Kan. Res. (pSB3K3) GFP) - K2560058
(Tet. Res. (pSB3T5) RFP) - K2560059
(Tet. Res. (pSB3T5) GFP) - K2560125 (Carb. Res. RFP)
- K2560126 (Carb. Res. GFP)
- K2560127 (Carb. Res. into BBa_K2560002)
- K2560132 (Cam. Res. into BBa_K2560002)
- K2560133
(Kan. Res. into BBa_K2560002) - K2560134
(Tet. Res. into BBa_K2560002)
Tags and Entry Vectors
| None |