Part:BBa_K2560014
Phytobrick version of BBa_J23106
This is the Phytobrick version of the promoter BBa_J23106 and was build as a part of the Marburg Collection. Instructions of how to use the Marburg Collection are provided at the bottom of the page.
Overview
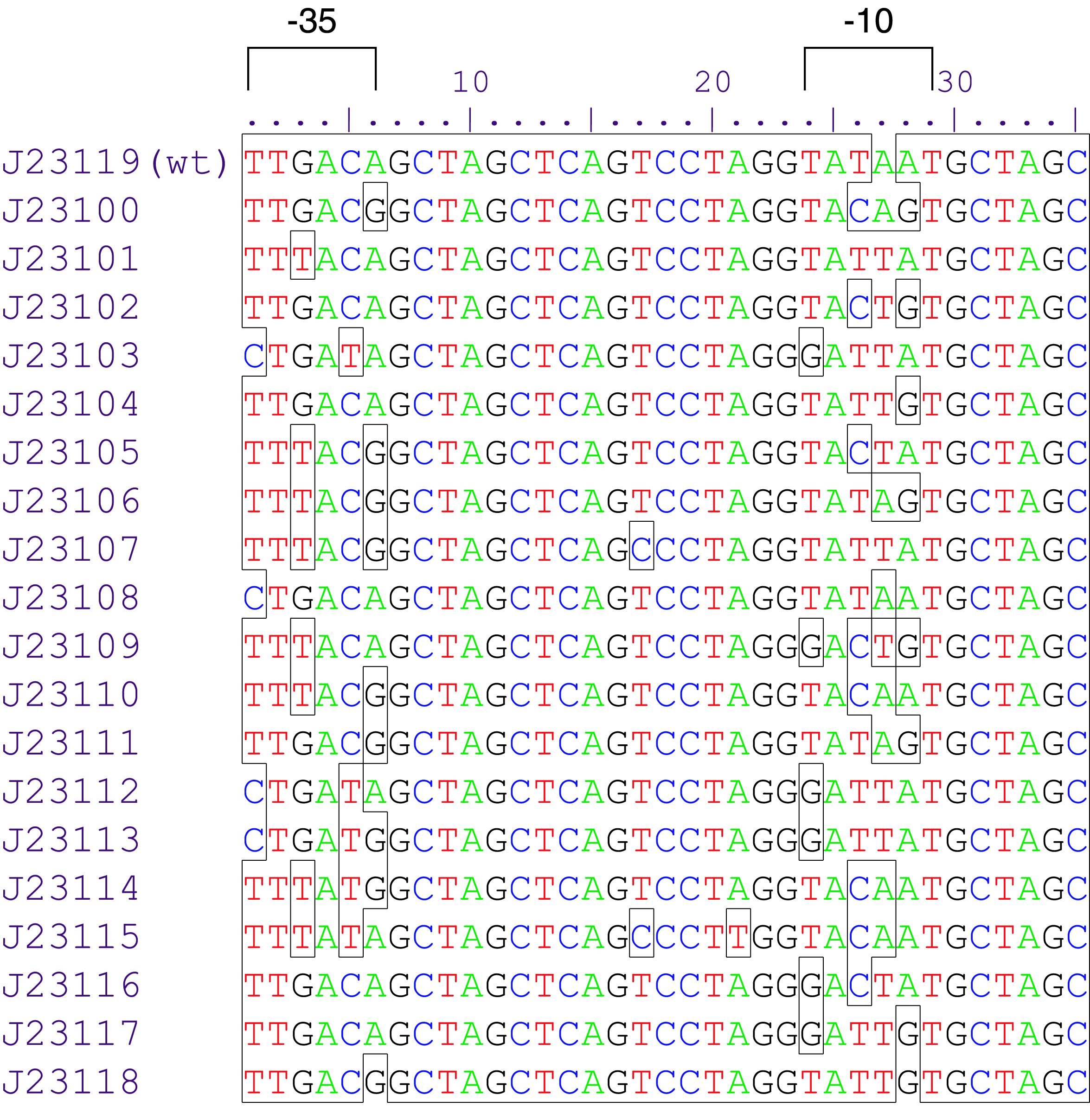
Promoters are genetic modules were the RNA polymerase is recruited to start RNA transcription. They are divided in two groups: constitutive promoters which transcribe RNA permanently and inducible promoters which start the transcription as a response to a stimulus. Inducible promoters can be regulated by transcription activation or repression. To start the RNA transcription the RNA polymerase complex is not sufficient. Therefore sigma70 factors are required. The sigma70 factor binds to the Pribnow box – two motifs -10 and -35bp upstream the CDS – recruiting the RNA transcrition complex enabling the transcription (Huerta and Collado, 2003), ( Paget and Helmann, 2003). Based on this knowledge a collection of synthetic constitutive promoters have been developed by Chris Anderson and made available on the iGEM repository.
Characterization
Variant Lux (au) K2560131 (Dummy) 0.025 K2560019 (J23103) 0.032 K2560026 (J23113) 0,038 K2560023 (J23109) 0,052 K2560009 (J23104) 0,058 K2560029 (J23117) 0,090 K2560025 (J23111) 0,098 K2560028 (J23116) 0,134 K2560021 (J23107) 0,136 K2560027 (J23114) 0,163 K2560024 (J23110) 0,169 K2560018 (J23102) 0,245 K2560030 (J23118) 0,348 K2560020 (J23105) 0,387 K2560015 (J23115) 0,398 K2560014 (J23106) 0,502 K2560017 (J23101) 0,510 K2560022 (J23108) 0,768 K2560007 (J23100) 1
We assembled 19 test plasmids with golden-gate-assembly and measured their expression strength, following our selfmade workflows. The results are shown in Figure 1. We observed an even distribution of the tested promoters throughout the dynamic range. The strongest promoter K2560007 (J23100) yielded 40 fold stronger signal than the promoter dummy and was used as a reference to calculate relative promoter strengths.
The test constructs were built with dummy connectors which did not possess insulator elements. We assume that this resulted in additional expression caused by transcription throughout the rest of the plasmid, e.g. ori and antibiotic resistance. This is thought to add the same extent of signal to all measured promoters thus reducing the overall dynamic range. To further evaluate this assumption, we could repeat this experiment with one of our insulators instead of the dummy connector.
Usage and Biology
Marburg 2018 characterized this part in Vibrio natriegens using the lux operon of Photorhabdus luminescens (BBa_K2560051). The parts sequence was verified by Sanger sequencing.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Marburg Toolbox
We proudly present the Marburg Collection, a novel golden-gate-based toolbox containing various parts that are compatible with the PhytoBrick system and MoClo. Compared to other bacterial toolboxes, the Marburg Collection shines with superior flexibility. We overcame the rigid paradigm of plasmid construction - thinking in fixed backbone and insert categories - by achieving complete de novo assembly of plasmids.
36 connectors facilitate flexible cloning of multigene constructs and even allow for the inversion of individual transcription units. Additionally, our connectors function as insulators to avoid undesired crosstalk.
The Marburg Collection contains 123 parts in total, including:
inducible promoters, reporters, fluorescence and epitope tags, oris, resistance cassettes and genome engineering tools. To increase the value of the Marburg Collection, we additionally provide detailed experimental characterization for V. natriegens and a supportive software. We aspire availability of our toolbox for future iGEM teams to empower accelerated progression in their ambitious projects.
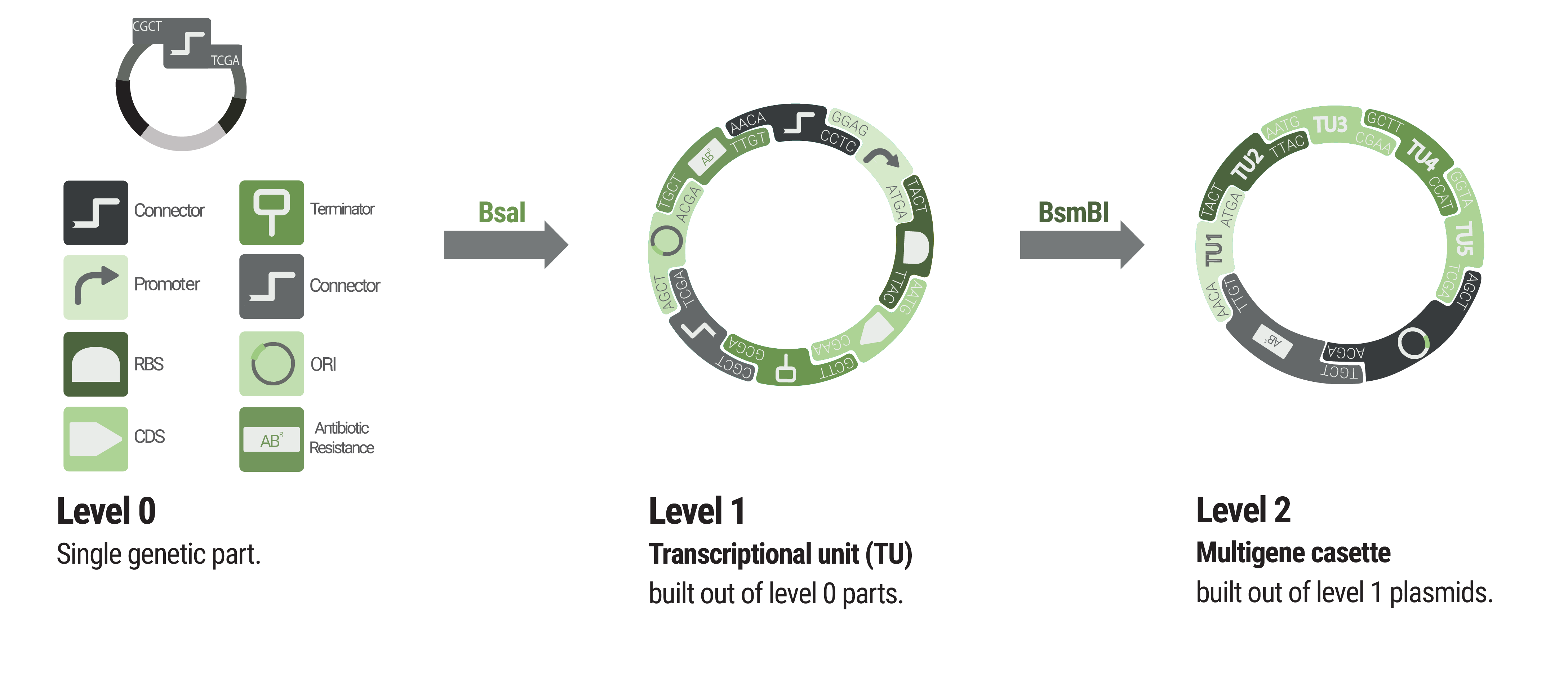
Basic building blocks like promoters or terminators are stored in level 0 plasmids. Parts from each category of our collection can be chosen to built level 1 plasmids harboring a single transcription unit. Up to five transcription units can be assembled into a level 2 plasmid.
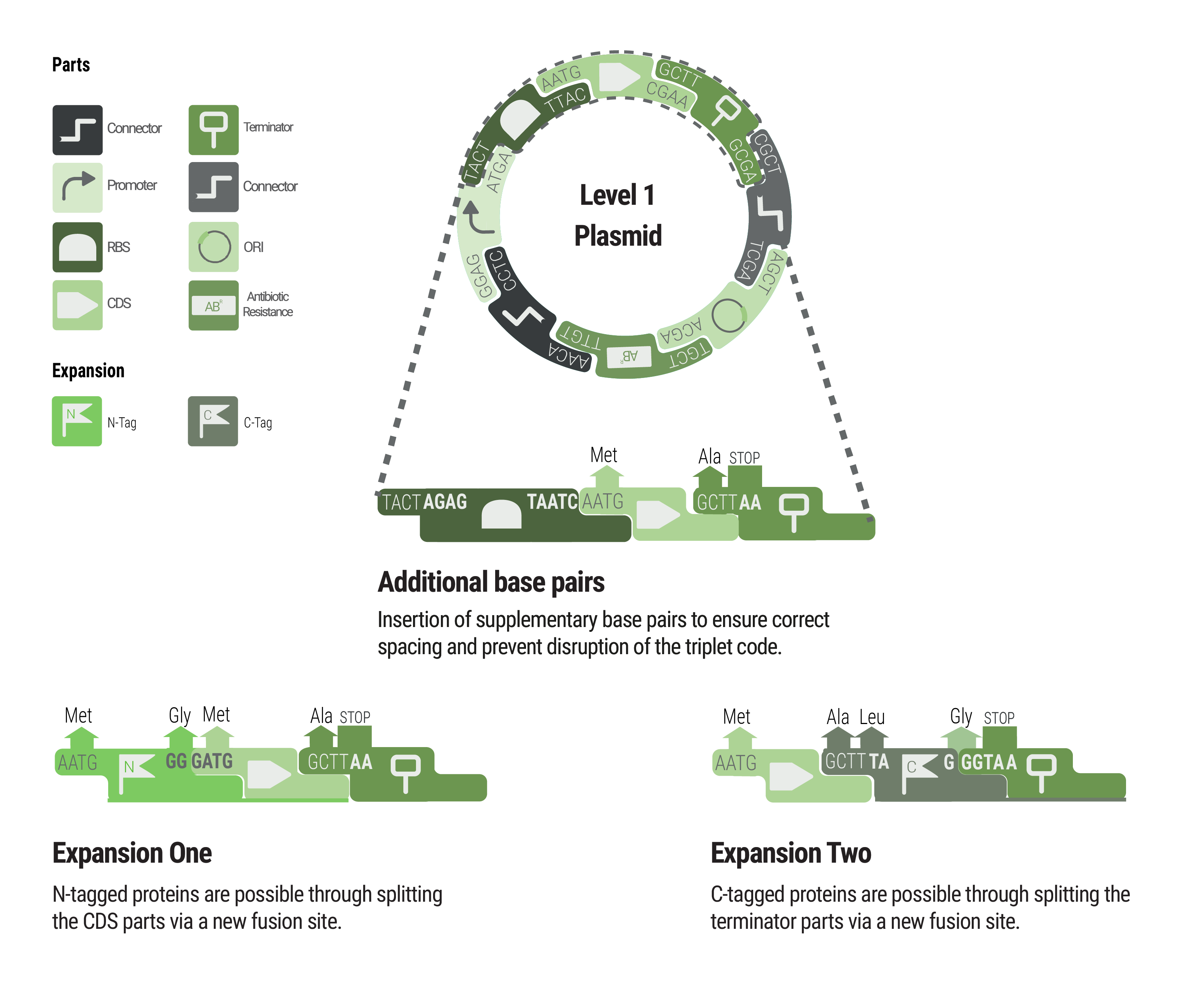
Between some parts, additional base pairs were integrated to ensure correct spacing and to maintain the triplet code. We expanded our toolbox by providing N- and C- terminal tags by creating novel fusions and splitting the CDS and terminator part, respectively.
Parts of the Marburg Toolbox

- K2560011 (5'Connector Dummy)
- K2560055
(1-6
Connector) - K2560065 (5'Con1)
- K2560066 (5'Con2)
- K2560067 (5'Con3)
- K2560068 (5'Con4)
- K2560069 (5'Con5)
- K2560075 (5'Con1
Short Res) - K2560076 (5'Con2
Short) - K2560077 (5'Con3
Short) - K2560078 (5'Con4
Short) - K2560079 (5'Con5
Short) - K2560095 (5'Con1 inv)
- K2560096 (5'Con2 inv)
- K2560097 (5'Con3 inv)
- K2560098 (5'Con4 inv)
- K2560099 (5'Con5 inv)
- K2560105 (5'Con5 inv
Ori) - K2560107 (5'Con1
Res)

- K2560007 (J23100)
- K2560009 (J23104)
- K2560014 (J23106)
- K2560015 (J23115)
- K2560017 (J23101)
- K2560018 (J23102)
- K2560019 (J23103)
- K2560020 (J23105)
- K2560021 (J23107)
- K2560022 (J23108)
- K2560023 (J23109)
- K2560024 (J23110)
- K2560025 (J23111)
- K2560026 (J23113)
- K2560027 (J23114)
- K2560028 (J23116)
- K2560029 (J23117)
- K2560030 (J23118)
- K2560031 (J23119)
- K2560123
(pTet) - K2560124 (pTrc)
- K2560131 (Promoter Dummy)

- K2560012 (3'Connector Dummy)
- K2560070 (3'Con1)
- K2560071 (3'Con2)
- K2560072 (3'Con3)
- K2560073 (3'Con4)
- K2560080 (3'Con5 Ori)
- K2560100 (3'Con1 inv
Short) - K2560101 (3'Con2 inv
Short) - K2560102 (3'Con3 inv
Short) - K2560103 (3'Con4 inv
Short) - K2560104 (3'Con5 inv
Short) - K2560106 (3'Con1 inv
Short Res) - K2560108 (3'Con1 inv)
- K2560109 (3'Con1 inv
Res) - K2560110 (3'Con2 inv)
- K2560111 (3'Con3 inv)
- K2560112 (3'Con4 inv)
- K2560113 (3'Con5 inv)

- K2560048 (Cam. Res. RFP)
- K2560056
(Kan. Res. (pSB3K3) RFP) - K2560057
(Kan. Res. (pSB3K3) GFP) - K2560058
(Tet. Res. (pSB3T5) RFP) - K2560059
(Tet. Res. (pSB3T5) GFP) - K2560125 (Carb. Res. RFP)
- K2560126 (Carb. Res. GFP)
- K2560127 (Carb. Res. into BBa_K2560002)
- K2560132 (Cam. Res. into BBa_K2560002)
- K2560133
(Kan. Res. into BBa_K2560002) - K2560134
(Tet. Res. into BBa_K2560002)
Tags and Entry Vectors
| None |