Part:BBa_K2560001
Phytobrick Entry Vector with RFP dropout
This plasmid can be used to create Phytobricks using a Golden Gate Reaction. A RFP dropout helps to distinguish between wrong colonies (red) and correct colonies (white)
Overview
All LVL0 parts have to be stored in plasmids to allow for amplification and long term storage. To create new LVL0 parts, a PCR product or annealed oligos are cloned into a part entry vector. This vector harbours the resistance and ori that are required for selection and propagation. Furthermore, part entry vectors can be designed in a way that they contain a dropout. This dropout can be a transcription unit for a marker that generates a visible output. The first golden-gate-based toolbox MoClo (Weber et al. 2011.) used a LacZ alpha transcription unit which can be used for blue white screening in many E. coli cloning strains. This concept was also adapted by iGEMs PhytoBrick system. During the cloning of LVL0 parts, this dropout is replaced by the desired part. When the cloning reaction is transformed into a suitable E. coli strain and the cells are plated on agar plates with supplemented IPTG and X-Gal. Colonies transformed with the religated entry plasmid appear blue while white colonies most probably contain the correctly assembled plasmid. The LVL0 part entry vector in iGEMs PhytoBrick system (BBa_P10500) has been designed as described and can be used for blue white screening.
We appreciate the approach of using part entry plasmids with dropouts but, for two reasons, we think that LacZ is not an optimal reporter. First, blue white screening requires the two expensive chemicals IPTG and X-Gal which have to be added to the agar plates. Second, blue white screening is restricted to E.coli strains with an incomplete lac operon that is complemented by the LacZ alpha fragment that is expressed from the plasmid (Langley et al. 1975.) . Consequently blue white screening is not compatible with a V. natriegens wild type strain (Link zu Improvement Page).
Experience
In our project we used this as a vector for using Golden Gate cloning to build our construct. The part was synthesized with added RFC10 prefix and suffix and then ligated to pSB1C3 according to the RFC10 standard. The vector plasmid was then purified and used in Golden Gate cloning. The new constructs made using this vector were then transformed into TOP10 cells which were grown on LB agar plates. The red fluorescence dropout colonies were easy to see by visual inspection but required over 24 hours after transformation for the RFP to be visible to the naked eye. The plates could be put in cold storage (4 °C) after 16 hours without any noticeable delay of the colonies becoming red [Aalto-Helsinki 2020].
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal SpeI site found at 51
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 21
Illegal NheI site found at 44
Illegal SpeI site found at 51 - 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal SpeI site found at 51
- 25INCOMPATIBLE WITH RFC[25]Illegal SpeI site found at 51
Illegal AgeI site found at 630
Illegal AgeI site found at 742 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1
Illegal BsaI.rc site found at 927
Marburg Toolbox
We proudly present the Marburg Collection, a novel golden-gate-based toolbox containing various parts that are compatible with the PhytoBrick system and MoClo. Compared to other bacterial toolboxes, the Marburg Collection shines with superior flexibility. We overcame the rigid paradigm of plasmid construction - thinking in fixed backbone and insert categories - by achieving complete de novo assembly of plasmids.
36 connectors facilitate flexible cloning of multigene constructs and even allow for the inversion of individual transcription units. Additionally, our connectors function as insulators to avoid undesired crosstalk.
The Marburg Collection contains 123 parts in total, including:
inducible promoters, reporters, fluorescence and epitope tags, oris, resistance cassettes and genome engineering tools. To increase the value of the Marburg Collection, we additionally provide detailed experimental characterization for V. natriegens and a supportive software. We aspire availability of our toolbox for future iGEM teams to empower accelerated progression in their ambitious projects.
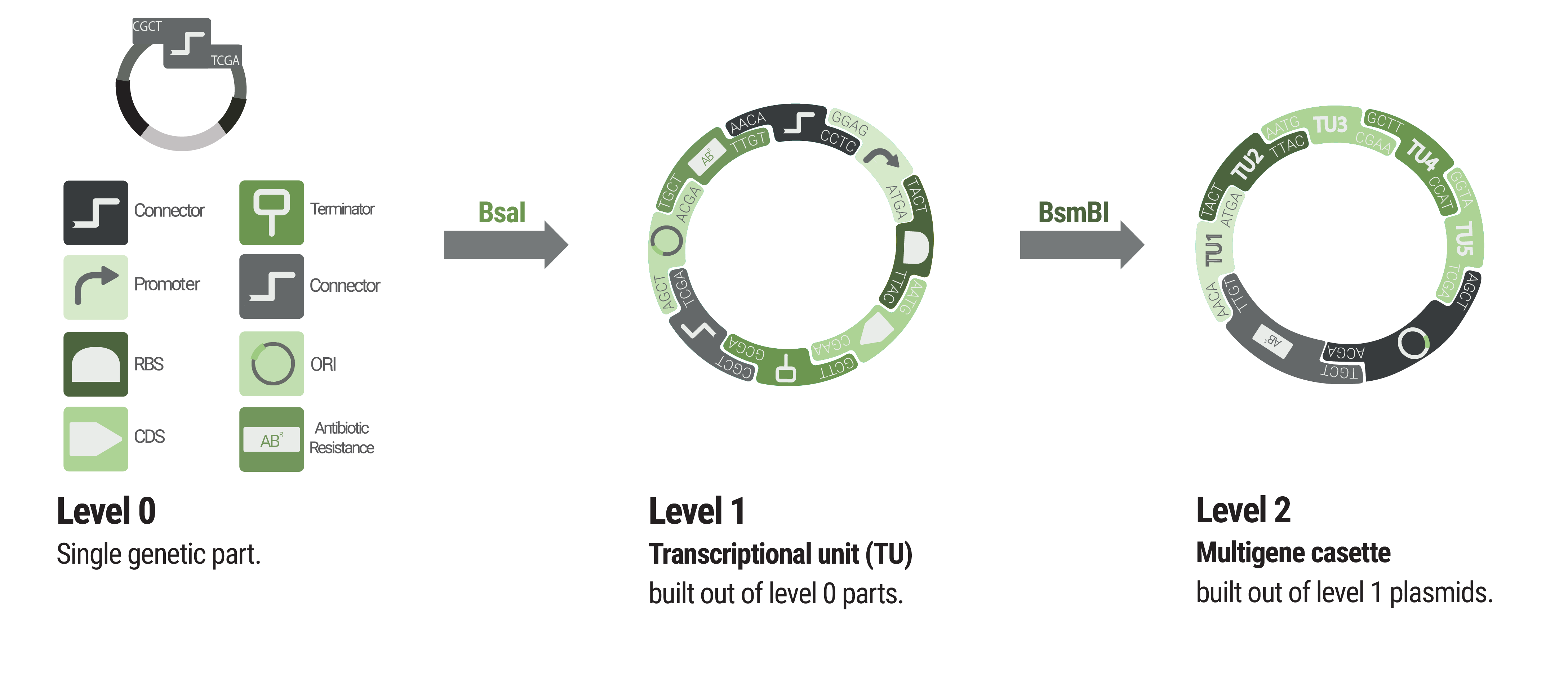
Basic building blocks like promoters or terminators are stored in level 0 plasmids. Parts from each category of our collection can be chosen to built level 1 plasmids harboring a single transcription unit. Up to five transcription units can be assembled into a level 2 plasmid.
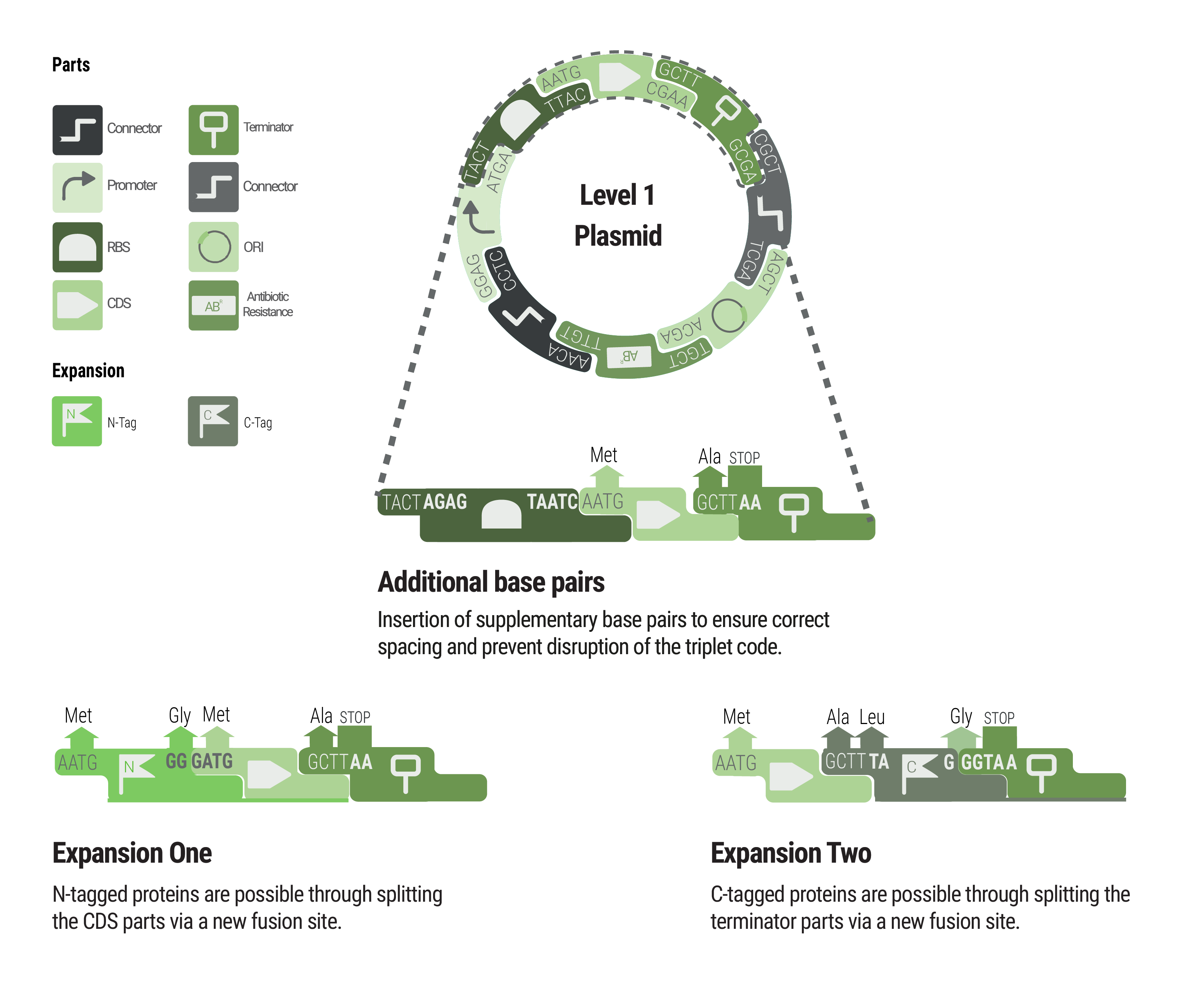
Between some parts, additional base pairs were integrated to ensure correct spacing and to maintain the triplet code. We expanded our toolbox by providing N- and C- terminal tags by creating novel fusions and splitting the CDS and terminator part, respectively.
Parts of the Marburg Toolbox

- K2560011 (5'Connector Dummy)
- K2560055
(1-6
Connector) - K2560065 (5'Con1)
- K2560066 (5'Con2)
- K2560067 (5'Con3)
- K2560068 (5'Con4)
- K2560069 (5'Con5)
- K2560075 (5'Con1
Short Res) - K2560076 (5'Con2
Short) - K2560077 (5'Con3
Short) - K2560078 (5'Con4
Short) - K2560079 (5'Con5
Short) - K2560095 (5'Con1 inv)
- K2560096 (5'Con2 inv)
- K2560097 (5'Con3 inv)
- K2560098 (5'Con4 inv)
- K2560099 (5'Con5 inv)
- K2560105 (5'Con5 inv
Ori) - K2560107 (5'Con1
Res)

- K2560007 (J23100)
- K2560009 (J23104)
- K2560014 (J23106)
- K2560015 (J23115)
- K2560017 (J23101)
- K2560018 (J23102)
- K2560019 (J23103)
- K2560020 (J23105)
- K2560021 (J23107)
- K2560022 (J23108)
- K2560023 (J23109)
- K2560024 (J23110)
- K2560025 (J23111)
- K2560026 (J23113)
- K2560027 (J23114)
- K2560028 (J23116)
- K2560029 (J23117)
- K2560030 (J23118)
- K2560031 (J23119)
- K2560123
(pTet) - K2560124 (pTrc)
- K2560131 (Promoter Dummy)

- K2560012 (3'Connector Dummy)
- K2560070 (3'Con1)
- K2560071 (3'Con2)
- K2560072 (3'Con3)
- K2560073 (3'Con4)
- K2560080 (3'Con5 Ori)
- K2560100 (3'Con1 inv
Short) - K2560101 (3'Con2 inv
Short) - K2560102 (3'Con3 inv
Short) - K2560103 (3'Con4 inv
Short) - K2560104 (3'Con5 inv
Short) - K2560106 (3'Con1 inv
Short Res) - K2560108 (3'Con1 inv)
- K2560109 (3'Con1 inv
Res) - K2560110 (3'Con2 inv)
- K2560111 (3'Con3 inv)
- K2560112 (3'Con4 inv)
- K2560113 (3'Con5 inv)

- K2560048 (Cam. Res. RFP)
- K2560056
(Kan. Res. (pSB3K3) RFP) - K2560057
(Kan. Res. (pSB3K3) GFP) - K2560058
(Tet. Res. (pSB3T5) RFP) - K2560059
(Tet. Res. (pSB3T5) GFP) - K2560125 (Carb. Res. RFP)
- K2560126 (Carb. Res. GFP)
- K2560127 (Carb. Res. into BBa_K2560002)
- K2560132 (Cam. Res. into BBa_K2560002)
- K2560133
(Kan. Res. into BBa_K2560002) - K2560134
(Tet. Res. into BBa_K2560002)
Tags and Entry Vectors
| None |







