Plasmid backbones/Construction/Parts
< Back to Plasmid Construction
| Part assembly | System operation | Protein expression | Assembly of protein fusions | Part measurement | Screening of part libraries | Building BioBrick vectors | DNA synthesis | Other standards | Archive |
| Or get some help on plasmid backbones. |
Here we include a catalog of all available parts that can compose plasmid backbones for you to browse. Or alternatively, use the links above to jump to a particular class of plasmid backbone parts.
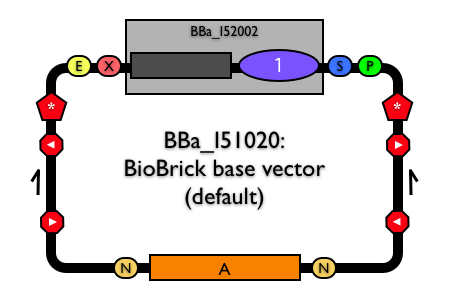
BioBrick base vector
View the BioBrick base vector BBa_I51020.
In designing a BioBrick base vector that will serve as the basis for many new vectors, our priority was to ensure that the resulting vectors would fulfill the basic demands of the biological engineers who use them. We had five design requirements for our new BioBrick vectors.
- BioBrick standard vectors must be able to propagate any BioBrick standard biological part. To meet this requirement, the base vector must contain a complete BioBrick cloning site as specified in the original technical standardKnight-2003.
- BioBrick standard vectors must be easy to use for their most common purpose: assembly of BioBrick standard biological parts. To meet this requirement, we included BBa_P1016 within the BioBrick cloning site. BBa_P1016 encodes the positive selection marker ccdB. Positive selection markers prevent one of the most common problems during assembly of BioBrick parts: contamination of the ligation reaction with uncut plasmid DNASambrook-2001. Any cells transformed with the uncut plasmid DNA produce the lethal protein ccdB and dieBernard-Gene-1994 Bernard-Gene-1995 Bernard-Biotechniques-1996. Note, however, that the drawback of this solution is that inclusion of this part requires that users propagate both the base vector and any derived vectors in E. coli strains tolerant of ccdB expression such as DB3.1Bernard-J-Mol-Biol-1992 Miki-J-Mol-Biol-1992.
- BioBrick standard vectors should be easy to purify. To meet this requirement, we included a pUC19-derived origin (BBa_I50022) in addition to the ccdB selection marker within the BioBrick cloning siteVieira-Gene-1982 Norrander-Gene-1983 Yanisch-Perron-Gene-1985. The high copy origin encoded by BBa_I50022 means that both base vector DNA and any derived vector DNA are easily purified in large quantities, irrespective of whether the vector replication origin is low copy or notCabello-Nature-1976 Ioannou-Nat-Genet-1994. Cloning a BioBrick part into the BioBrick cloning site removes the high copy origin in the cloning site thereby restoring replication control to the vector origin.
- BioBrick standard vectors must stably propagate BioBrick parts. Previous work suggests that transcriptional isolation of the cloned DNA fragment can make those fragments more amenable to manipulationGentz-Proc-Natl-Acad-Sci-USA-1981 Stueber-EMBO-J-1982 Bowater-Nucleic-Acids-Res-1997 Godiska-2005. Thus, to meet this requirement, we included transcriptional terminators in both directions flanking the BioBrick cloning site (BBa_B0053, BBa_B0054, BBa_B0055 and BBa_B0062). By doing so, we sought to insulate the proper maintenance and propagation of the vector from the possibly disruptive function encoded by the BioBrick part cloned into the vector. We also flanked the BioBrick cloning site on both sides with translational stop codons in all six reading frames (BBa_B0042) to ensure translational insulation as well.
- BioBrick standard vectors should allow users to verify the length and sequence of the inserted BioBrick part. To meet this requirement and to support backwards compatibility with existing BioBrick parts and vectors, we incorporated the same primer annealing sites found in existing BioBrick vectors like pSB1A3-P1010 (BBa_G00100 and BBa_G00102). BBa_G00100 and BBa_G00102 are sufficiently distant from the BioBrick cloning site to ensure good quality sequence reads of any inserted part.
BBa_I51020 (BioBrick base vector) is composed of the BioBrick parts described. The BioBrick vector parts developed in this work confer upon the base vector, and therefore also any derived BioBrick standard vectors, the features that meet our design requirements.
To ensure that BBa_I51020 (BioBrick base vector) is easy to propagate and purify, we made BBa_I51020 itself a functional plasmid. We included an ampicillin resistance marker BBa_P1006 flanked by NheI restriction endonuclease sites (BBa_B0045) in the base vector BBa_I51020. BBa_P1006 is excised from BBa_I51020 upon construction of a new BioBrick standard vector. Thus, the combination of BBa_I50022 (high-copy replication origin) in the BioBrick cloning site and BBa_P1006 renders BBa_I51020 capable of autonomous plasmid replication for easy DNA propagation and purification.
Replication origins
The genetic element responsible for the replication of plasmids during cell growth and division is called a replication origin (also "origin of replication" or simply "origin"). There are several different replication origins and they differ in their plasmid copy number per cell (how many molecules of the plasmid are maintained in the cell), mechanism of copy number control, cell-to-cell copy number variation, and even the degree of coiling of the physical DNA. Thus, BioBrick® parts, devices and systems can operate very differently from one plasmid backbone to another.
| Several replication origins have been designed in the Registry. Although most of them are not available as individual BioBrick parts, some can be obtained by PCR of existing plasmid backbones in the Registry using appropriate primers. In fact, when making new BioBrick vectors using the BioBrick base vector, Reshma simply assembled a PCR-amplified linear DNA encoding the replication origins with the antibiotic resistance markers. (The PCR primers included the BioBrick prefix and suffix so that the linear DNA encoding the origin was ready for digestion with EcoRI and SpeI. |
| Name | Description | Replicon | Copy number | Chassis | Length |
|---|---|---|---|---|---|
| BBa_I50000 | F plasmid backbone with BioBrick sites removed. | 4640 | |||
| BBa_I50001 | F plasmid backbone with BioBrick sites removed, reverse | 4640 | |||
| BBa_I50022 | Minimal pUC19-derived high copy replication origin | 674 | |||
| BBa_I50030 | pBR322 replication origin | 2016 | |||
| BBa_I50032 | p15A replication origin | 1554 | |||
| BBa_I50040 | nonfunctional pSC101 origin of replication | 2226 | |||
| BBa_I50041 | pSC101 origin of replication, reverse | 2037 | |||
| BBa_I50042 | pSC101 origin of replication | 2037 | |||
| BBa_I50050 | R6K replication origin | 312 | |||
| BBa_I50052 | pSC101 temperature sensitive origin of replication | 2037 | |||
| BBa_J61001 | [R6K] Origin of replication | 406 | |||
| BBa_K112125 | OriV origin of replication | 646 | |||
| BBa_K125020 | lac promoter/rbs/rep/oriV/lac promoter/rbs/aadA/oriT | 4841 | |||
| BBa_K125340 | Origin of Vegetative Replication | 424 | |||
| BBa_K320000 | Asaia replication origin | 1391 | |||
| BBa_K320004 | Origin for Asaia and resistance to Kanamycin | 2374 | |||
| BBa_K524203 | pSC101 heat sensitive origin of replication with deletion in par locus | 1941 | |||
| BBa_K533000 | p15A replication origin | relaxed, high copy number | 914 | ||
| BBa_K864050 | pSC101 low copy origin of replication | 2216 | |||
| BBa_K864051 | pSC101ts temperature sensitive low copy origin of replication | 2216 |
Antibiotic resistance markers
| At the most basic level, function of the antibiotic resistance marker is to allow the cell to grow even in the presence of a particular antibiotic. Plasmid backbones include antibiotic resistance markers because the markers allow you to select for cells that contain your plasmid. When E. coli cells grow and divide, plasmids can inadvertently be lost from the cell. In some cases, cells without a plasmid can potentially grow faster than cells with the plasmid which means that cell cultures can quickly become dominated by plasmid-free cells. Since most BioBrick® parts are maintained on plasmid backbones, plasmid loss is problematic. To help avoid these problems, every plasmid backbone includes an antibiotic resistance marker. Thus, cells which don't have a copy of the plasmid are killed by antibiotic present. Common antibiotic resistance markers in BioBrick® plasmid backbones confer resistance to ampicillin ("Amp" or A), kanamycin ("Kan" or K), chloramphenicol ("Cm" or C) and tetracycline ("Tet" or T). |
Several common antibiotic resistance markers are available from the Registry. These markers include a promoter, ribosome binding site, and coding sequence for the protein or enzyme that confers resistance to the antibiotic. Thus, inclusion of one of these markers in a new BioBrick plasmid is sufficient to confer antibiotic resistance upon cells that maintain that plasmid.
| Name | Description | Resistance | Length |
|---|---|---|---|
| BBa_P1002 | ampicillin resistance cassette | 943 | |
| BBa_P1004 | chloramphenicol resistance cassette | 769 | |
| BBa_P1000 | chloramphenicol resistance cassette | 792 | |
| BBa_J61000 | chloramphenicol resistance cassette | 894 | |
| BBa_K292004 | Double terminator + Kanamycin resistance cassette + Double ter | 1173 | |
| BBa_K4047016 | Gentamicin resistance (GmR) for Synechococcus | 534 | |
| BBa_P1014 | gentamicin resistance cassette aph(2) | Gentamicin 25 ug/ml | 1089 |
| BBa_P1013 | gentamycin resistance cassette (aacC1 gene) | 841 | |
| BBa_P1012 | gentamycin/kanamycin resistance cassette | 1943 | |
| BBa_P1003 | kanamycin resistance cassette | 967 | |
| BBa_K292003 | Kanamycin resistance cassette and a double terminator | 1070 | |
| BBa_K1642003 | Kanamycin Resistance Cassette in Cyanobacteria | 1405 | |
| BBa_K3521007 | Kanamycin Resistance Cassette(KanR) | 813 | |
| BBa_K1937002 | OriKan: a composite part to turn any pSB1C3 into a Bacillus plasmid | 2489 | |
| BBa_K4047027 | SmR | 792 | |
| BBa_P1001 | tetracycline resistance cassette | 1279 | |
| BBa_P1005 | tetracycline resistance cassette | 1283 | |
| BBa_K1033990 | Zeocin resistance cassette | E coli: 25 ug/ml zeocin | 449 |
| Chris Anderson, a professor of bioengineering at UC Berkeley, constructed the antibiotic resistance marker BBa_J61000. | Austin Che constructed the antibiotic resistance markers BBa_P1000 and BBa_P1001 in Tom Knight's lab. | ||
| Tom Knight constructed the antibiotic resistance marker BBa_P1014. |
Transcriptional and translational insulation parts
Ideally, the operation of the plasmid backbone should be insulated from the operation of the cloned BioBrick part, device or system. For example, transcriptional and translational insulation can make it easier to clone and propagate some BioBrick parts, devices, or systems that are toxic to the cellular chassis when transcribed or expressed. In other cases, the proper function of a BioBrick part, device, or system may depend on there being no basal transcription or translation of the cloned component.
| To transcriptionally insulate the plasmid backbone from the cloned BioBrick part in new BioBrick vectors constructed from the base vector, Reshma designed two forward and two reverse transcriptional terminators to flank the BioBrick cloning site and prevent transcriptional read-through into or out of the cloning site. Note, that although some of these terminators have been used previously in plasmid backbones, that their transcriptional termination efficiency has not yet been measured. (Measuring their transcriptional efficiency would be a great project for an iGEM team or student interested in synthetic biology!) |
| To translationally insulate the plasmid backbone from the cloned BioBrick part in new BioBrick vectors constructed from the base vector, Reshma flanked the BioBrick cloning site with a translational stop sequence previously designed by Randy Rettberg. The translational stop sequence includes stop codons in all reading frames on both strands and thus should prevent translational read-through into or out of the cloning site. |
| Name | Description | Direction | Efficiency Fwd. Rev. | Chassis | Length | |
|---|---|---|---|---|---|---|
| BBa_B0042 | Translational stop sequence | 12 | ||||
| BBa_B0053 | Terminator (His) | Forward | 72 | |||
| BBa_B0054 | Transcriptional terminator | 69 | ||||
| BBa_B0055 | -- No description -- | 78 | ||||
| BBa_B0062 | Terminator (Reverse B0052) | Reverse | 41 | |||
Primer binding sites
| A common operation with BioBrick plasmids is to verify the length or sequence of the cloned BioBrick part. To verify the length of the BioBrick part, most people use [http://openwetware.org/wiki/Colony_PCR colony PCR] to amplify the BioBrick part and check the length using [http://openwetware.org/wiki/Agarose_gel_electrophoresis agarose gel electrophoresis]. To [http://openwetware.org/wiki/Sequencing_DNA sequence the part], most people use a core facility at their institution or send samples to commercial DNA sequencing services. Regardless, both length and sequence verification require the use of primers (short synthetic DNA fragments usually ~20 nucleotides in length). For convenience, most BioBrick plasmid backbones, including those constructed from the BioBrick base vector, include standard primer annealing sites for the VF2 and VR primers. The VF2 and VR primers flank the BioBrick cloning site but are sufficiently distant from the cloning site to ensure high quality sequencing reads. Or alternatively, you can also use the BioBrick-f and BioBrick-r primers which anneal to the BioBrick prefix and BioBrick suffix sequences, respectively. | |
Note that the VF2 and VR primers do not work well with all BioBrick parts. See notes on using the VF2 and VR primers.
| Name | Description | Sequence | Length |
|---|---|---|---|
| BBa_G00000 | BioBrick cloning site prefix | gaattcgcggccgcttctagag | 22 |
| BBa_G00001 | BioBrick cloning site suffix | tactagtagcggccgctgcag | 21 |
| BBa_G00100 | Forward primer for sequencing/amplifying BioBrick parts (VF2) | tgccacctgacgtctaagaa | 20 |
| BBa_G00102 | Reverse BioBrick primer annealing site (VR binding site) | gctcactcaaaggcggtaat | 20 |
Cloning sites
| To adhere to the BioBrick standard for physical composition of genetic parts, plasmid backbones must include a BioBrick cloning site. The cloning site includes a BioBrick prefix sequence composed of an EcoRI, NotI, and XbaI restriction enzyme recognition site and a BioBrick suffix sequence composed of a SpeI, NotI, and PstI restriction enzyme recognition site. | |
| In most BioBrick plasmid backbones, including those constructed from the BioBrick base vector, the BioBrick prefix and suffix sequences are not adjacent to one another. Instead, the BioBrick prefix and suffix flank what is a called a default plasmid insert or default insert for short. There are several different default inserts available in the Registry. See default plasmid inserts for more information. |
| Name | Description | Sequence | Length |
|---|---|---|---|
| BBa_G00000 | BioBrick cloning site prefix | gaattcgcggccgcttctagag | 22 |
| BBa_G00001 | BioBrick cloning site suffix | tactagtagcggccgctgcag | 21 |
Default plasmid inserts
Plasmid backbones available from the Registry come with a default part or plasmid insert in the BioBrick cloning site. There are a handful of default plasmid inserts used, as listed below.
Most of the default inserts used (including those constructed from the BioBrick base vector) encode the [http://openwetware.org/wiki/CcdB ccdB gene]. The ccdB gene encodes the toxic protein CcdB. The ccdB gene is as a positive selection marker during assembly or cloning of BioBrick parts into a BioBrick plasmid. Any BioBrick plasmid backbones that either are not cut during the restriction digest or re-ligate to the default insert will express the toxic protein CcdB upon transformation and kill the cell.
The default plasmid insert BBa_I52002 contains not only the positive selection marker ccdB but also a high copy replication origin. BBa_I52002 is used in BioBrick plasmids that contain a low or medium copy replication origin. The high copy replication origin in the BioBrick cloning site ensures that the plasmid propagates at high copy and it is easy to purify plasmid DNA. When a new BioBrick part is cloned into the cloning site, the high copy origin is replaced and plasmid replication reverts to control by the low or medium copy origin.
| Name | Description | Length |
|---|---|---|
| BBa_I739400 | Default insert of plasmid BBa_I739201, BBa_I739202, BBa_I739204 | 9 |
| BBa_J23025 | OnRFP | 923 |
| BBa_K106400 | Default insert for AarI AD acceptor vectors | 52 |
Miscellaneous plasmid components
For completeness, we list additional plasmid components documented in the Registry that do not fall into one of the above categories. Please click on the part link for additional information.
| Name | Description | Sequence | Length |
|---|---|---|---|
| BBa_B0043 | Topoisomerase I mediated cloning site (forward) | cccttagtgactc | 13 |
| BBa_B0044 | Topoisomerase I mediated cloning site (reverse) | gagtcactaaggg | 13 |
| BBa_B0045 | NheI restriction enzyme site (XbaI, SpeI compatible overhang) | gctagc | 6 |
| BBa_K558005 | Zymomonas mobilis glucose facilitator gene | . . . cagattattaatccggcttttttattattt | 1564 |
| BBa_K653001 | GFP reporter plus Double Terminator | . . . caccttcgggtgggcctttctgcgtttata | 857 |
| BBa_K863201 | 3' UTR site of alcohol oxidase 1 gene (aox1) | . . . tcatcaacttgaggggcactatcttgtttt | 676 |
References
Replication origins
<biblio>
- Chang-JBacteriol-1978 pmid=149110
- Cohen-JBacteriol-1977 pmid=334752
- Bolivar-Gene-1977 pmid=344137
- Bazaral-J-Mol-Biol-1968 pmid=4939624
- Hershfield-Proc-Natl-Acad-Sci-USA-1974 pmid=4610576
</biblio>
Antibiotic resistance markers
<biblio>
- Watanabe-J-Bacteriol-1961 pmid=13783344
- Watanabe-Bacteriol-Rev-2003 pmid=13999115
- Anderson-Br-Med-J-1965 pmid=5321597
</biblio>