Plasmid backbones/Construction
| Part assembly | System operation | Protein expression | Assembly of protein fusions | Part measurement | Screening of part libraries | Building BioBrick vectors | DNA synthesis | Other standards | Archive |
| Or get some help on plasmid backbones. |
You may find that there does not already exist a BioBrick® plasmid backbone that fulfills your requirements. You may need a different antibiotic resistance, a different copy number, or something else.
For convenience, we also describe the approach here. Or, you may wish to jump directly to the catalog of BioBrick parts for building BioBrick vectors.
Overview
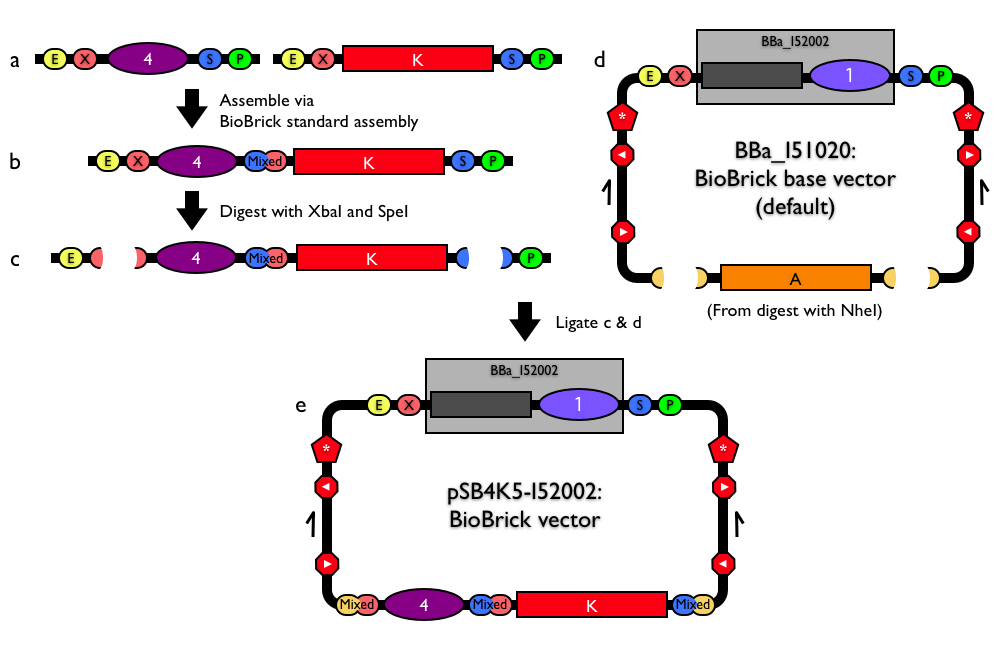
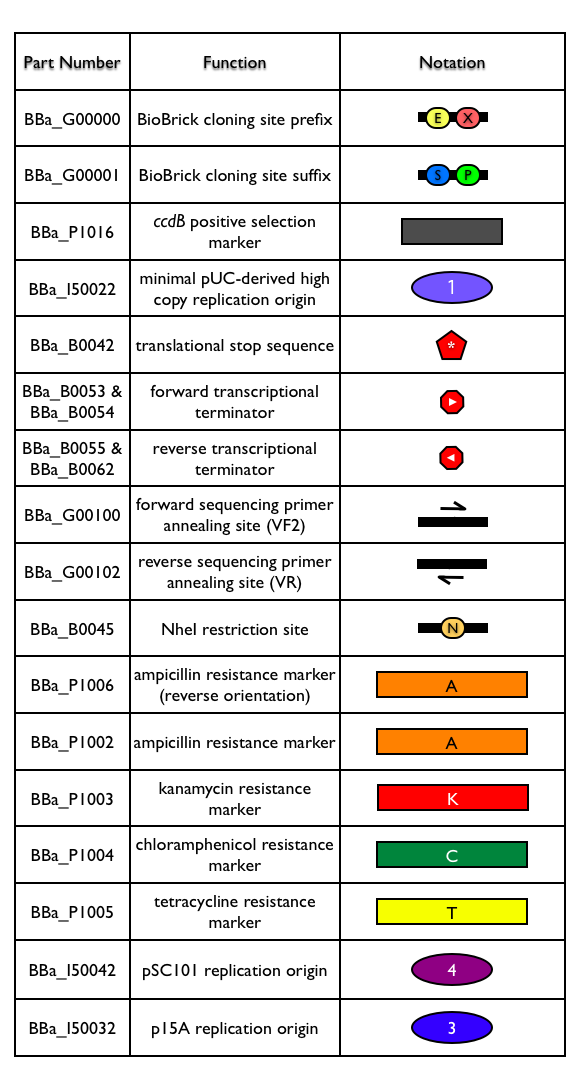
| Reshma's method for building new BioBrick® vectors relies on building BioBrick® vectors entirely from BioBrick® parts. To make the process of building BioBrick® vectors easier, Reshma designed a BioBrick® base vector (BBa_I51020). Using the based vector, you can construct a new BioBrick® vector from (almost) any replication origin and antibiotic resistance marker, as long as the origin and antibiotic resistance marker are themselves BioBrick® parts. In a nutshell, the method allows you to assembly any replication origin and antibiotic resistance marker together into the BioBrick base vector to construct a new, functional BioBrick vector. The base vector already has many of the general plasmid features that you might need in a BioBrick® vector, so use of the base vector ensures standardization and uniformity of any resulting BioBrick® vectors. (A complete list of all design features of the BioBrick® base vector is available.) | 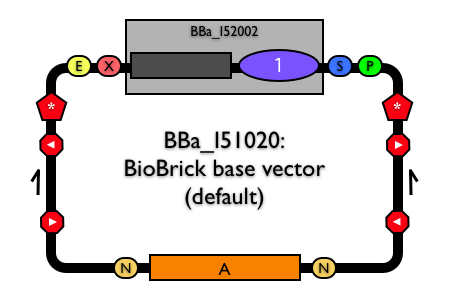
|
Method
Constructing any new BioBrick® standard construction plasmid starting from the BioBrick® base vector requires just two assembly steps.
|
This plasmid construction approach enables reuse of replication origins and antibiotic resistance markers available from the Registry of Standard Biological Parts. Use of the base vector to construct all BioBrick® standard construction plasmids ensures standardization and uniformity in any resulting BioBrick® vectors.
Entering new BioBrick plasmid backbones into the Registry
Please see the help page on entering new plasmid backbones and their default plasmid inserts into the Registry for details.
BioBrick plasmid parts
Browse all BioBrick plasmid parts.