Part:BBa_I50022
Minimal pUC19-derived high copy replication origin
|
BBa_I50022 is a minimal high-copy replication origin derived from pUC19. Any vector containing BBa_I50022 will be propagated at high copy. |
Usage and Biology
- [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=169656098 GenBank EU496091]
Characterization of ori pMB1 by iGEM team Aachen 2019
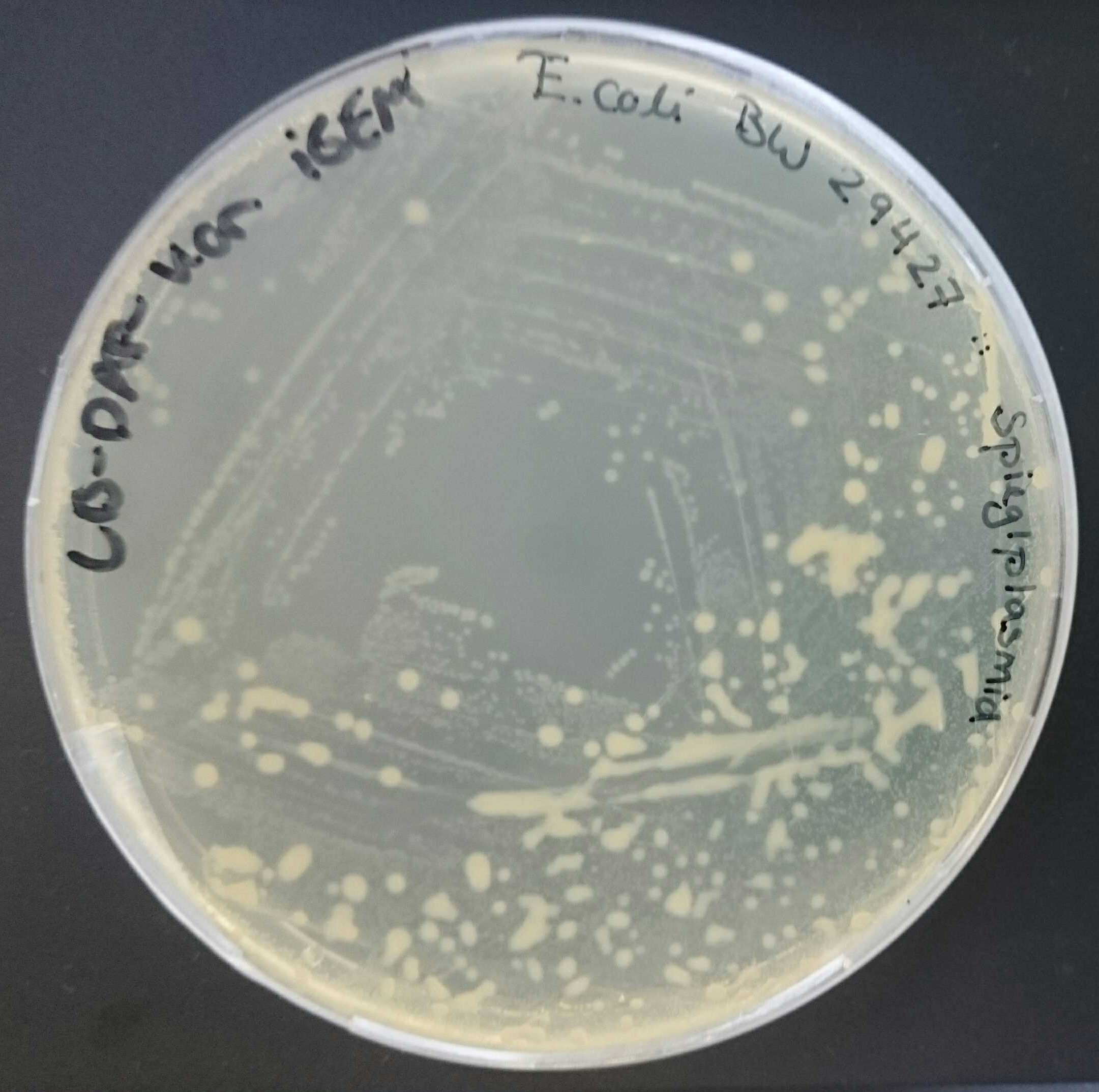
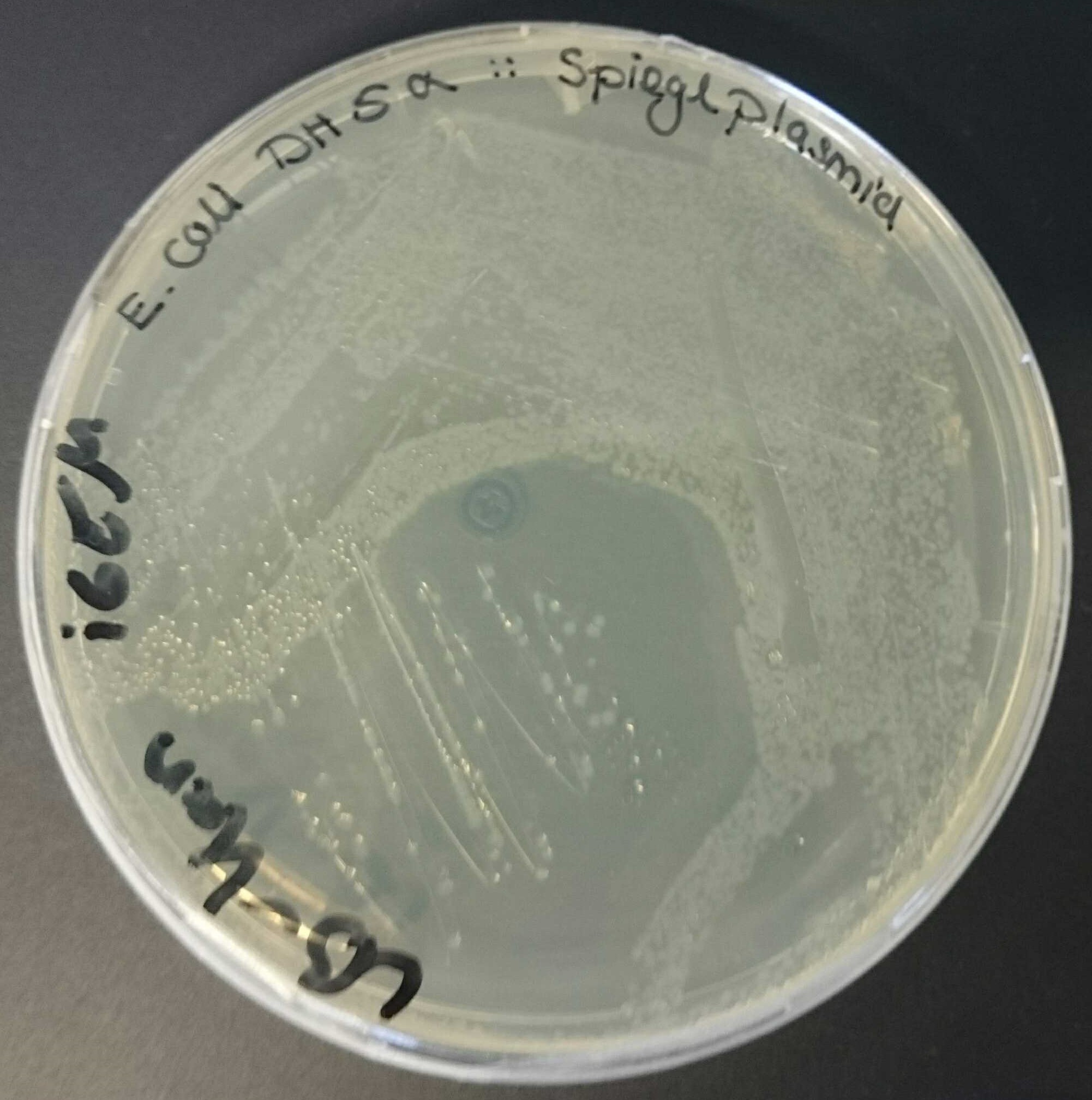
In our project we worked with the vector pBam containing ori R6K [pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. Martinez-Garcia E, Calles B, Arevalo-Rodriguez M, de Lorenzo V. BMC Microbiol. 2011 Feb 22;11:38. doi: 10.1186/1471-2180-11-38. 10.1186/1471-2180-11-38 PubMed 21342504]. As this ori has a low copy number [Morgan, K. (2014, February 14). Plasmids 101: Origin of Replication. Retrieved from https://blog.addgene.org/plasmid-101-origin-of-replication] it was difficult to get decent DNA concentrations after a plasmid prep (see also our characterization of BBa_J61001). So, we used ori pMB1 for amplification in E. coli DH5α. Therefore BBa_I50022 was cloned into pBam via restriction and ligation. Afterwards the new plasmid containing oriR6K and oripMB1 was transformed in two different chemically competent E. coli strains [Sambrook J, Fritsch EF, and Maniatis T (1989), Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.]. We’ve chosen E. coli DH5α as a strain to replicate plasmids containing ori pMB1 and E. coli BW29427 as a strain known to replicate plasmids containing ori R6K [Ye L., Hildebrand F., Dingemans J., Ballet S., Laus G., Matthijs S., … Cornelis P. (2014). Draft genome sequence analysis of a Pseudomonas putida W15Oct28 strain with antagonistic activity to Gram‐positive and Pseudomonas sp. pathogens. PLoS ONE, 9, e110038.]. As expected, both strains were able to grow on LB-Kan plates, as pBam contains a resistance gene for kanamycin (see figure 1 and figure 2).
To get data for the strength of the oris we prepped plasmids from both transformed strains. Therefore, we used a commercial kit (Nucleospin Macherey-Nagel) and normed the OD(600) of the used cultures to 2,55. 2 mL of culture was used per plasmid preparation. The resulting DNA concentrations are shown in table 1.
Table 1: DNA concentration after plasmid preparation of a plasmid containing ori R6K and ori pMB1 of different strains of E. coli
| Culture | DNA concentration [ng/μL] | Culture | DNA concentration [ng/μL] |
|---|---|---|---|
| E. coli DH5α 1 | 224,0 | E. coli BW29427 1 | 26,5 |
| E. coli DH5α 2 | 194,2 | E. coli BW29427 2 | 14,3 |
| E. coli DH5α 3 | 280,4 | E. coli BW29427 3 | 10,4 |
| E. coli DH5α 4 | 248,3 | E. coli BW29427 4 | 14,1 |
This data suggests that E. coli BW29427 is not able to use ori pMB1 as the resulting DNA concentrations do not differ from a plasmid preparation with E. coli BW29427 carrying a plasmid with ori R6K without ori pMB1 (table 2).
Table 2: DNA concentrations [ng/μL] after plasmid preparation of transformed E. coli BW29427 with pBam containing only oriR6K as a n origin of replication.
| Culture number | DNA concentration [ng/μL] |
|---|---|
| 1 | 12,4 |
| 2 | 14,6 |
| 3 | 13,9 |
| 4 | 14,5 |
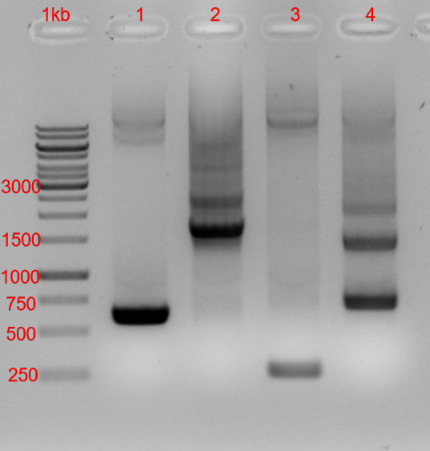
E. coli DH5α can amplify the transformed plasmid as expected. We also transformed E. coli DH5α with pBam containing only ori R6K as an origin of replication, but there was no growth on LB-Kan plates (the same protocol and cells from the same batch of chemically competent cells was used). A PCR was done to show the presence of ori R6K and ori pMB1 in the purified plasmid from E. coli DH5α. We even checked the direction of ori pMB1 in the plasmid. A picture of the resulting gel is shown below (figure 3).
Figure 3: 1,2 % agarose gel (150 V, 50 W, 300 mA, 25 min) of plasmid preparation from E. coli DH5α. The numbers indicate different primer combinations. 1: primers for ori R6K forward (620 bp expected), 2: primers for ori pMB1 reverse (1600 bp expected), 3: primers for ori pMB1 forward (1650 bp expected), 4: ori pMB1 forward and ori pMB1 reverse primer (700 bp expected)
This gel (figure 2) indicates that ori R6K (in forward direction) and ori pMB1 (in reverse direction) are inside the plasmid. Through the expected band in line 4 the binding of the ori pMB1 forward primer is shown. This proves, that ori pMB1 is inserted in reverse direction. Other bands in line 2 and 4 could come from unspecific binding of the primers or binding in residual genomic DNA after the plasmid preparation. In conclusion we have proven that E. coli Dh5α can’t use ori R6K but ori pMB1 and E. coli BW29427 can use ori R6K and does not profit from ori pMB1. Also, we characterized the strength of those origins of replications by analyzing the plasmid yield after a plasmid preparation.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 19
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |