Part:BBa_J61001
[R6K] Origin of replication
R6K origin of replication requires a pir+ or pir116 strain for replication in the absence of a second origin. In most E. coli strains, this is a silent feature.
This part belongs to a family useful for the construction of transposons and knockin cassettes.
FRT The FRT sequence is the recombination substrate for Flp recombinase. Like the Cre/Lox system, sequences flanked by FRT sites oriented in the same direction get excised by expression of Flp. Cassettes flanked by FRT allow the introduction of selectable markers into the genome followed by subsequent removal by the recombinase.
Tn5 The two Tn5 elements are the substrates for the Tnp transposase in both orientations. Sequences flanked by the two Tn5 elements (in opposing orientation) are mobilizable elements. In the presence of Tnp, they are excised from their plasmid and introduced randomly into the genome or plasmids present in the cell.
Tnp The gene for a high-activity mutant of Tn5 transposase.
OriTr Special strains of E. coli with remnants of the RP4 plasmid, including Rlambda (part ) and BW20767, can transfer plasmids with oriTr (part J01003) to diverse species of bacteria.
OriTf Special strains of E. coli with remanants of the F plasmid, such as J23055, can transfer plasmids with oriTf (part J01002) to other E. coli.
R6K A conditional origin of transfer requiring the pir gene expressed in trans.
Construction of Transposon Plasmids
Transposition mutagenesis is usually carried out by mating a donor strain containing a transposon on a plasmid into a recipient strain. The plasmid with the transposon and transposase transfers to the recipient strain, the transposon is excised from the plasmid, and inserts randomly into the genome. A selectable marker in the transposon and in the recipient strain allows selection of cells receiving the transposon over both the donor and recipient strain.
Construction of the transposon donor strain requires a conjugative donor strain harboring a plasmid with the following elements:
- A conjugative origin of transfer
- A conditional origin of replication, usually R6K, so that the plasmids cannot replicate within the recipient strain (so no second origin of replication)
- A transposon containing a selectable marker flanked by Tn5 ends
- The transposase gene
Usage and Biology
Characterization by iGEM team Aachen 2019.
To characterize this origin of replication (ori) we tested it in two different strains of E. coli: E. coli BW 295427 (also known as WM3064) containing the pir gene and E. coli DH5α which does not contain the pri gene [Ye L., Hildebrand F., Dingemans J., Ballet S., Laus G., Matthijs S., … Cornelis P. (2014). Draft genome sequence analysis of a Pseudomonas putida W15Oct28 strain with antagonistic activity to Gram‐positive and Pseudomonas sp. pathogens. PLoS ONE, 9, e110038.]. To somehow quantify the strength of the ori in those strains, we transformed them with a plasmid containing ori R6K (pBam∷mamC-eGFP) [pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. Martinez-Garcia E, Calles B, Arevalo-Rodriguez M, de Lorenzo V. BMC Microbiol. 2011 Feb 22;11:38. doi: 10.1186/1471-2180-11-38. 10.1186/1471-2180-11-38 PubMed 21342504]and measured the DNA concentration after plasmid preparation with a commercial kit (Nucleospin Macherey-Nagel).
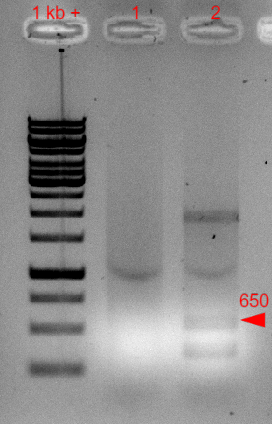
To choose a correct plasmid for transformation, we made a PCR of two prepped plasmids that should contain ori R6K.
The resulting picture of an 1,2 % agaraose gel of the PCR product is shown in fig.1 below.
Figure 1: 1,2 % agarose gel (120 V, 50 W, 300 mA, 25 min) of PCR from template plasmid used for transformation of E. coli BW29427 and E. coli DH5α. 1 and 2 are from two different plasmid preparations which should contain oriR6K. The additional bands in sample 2 could come from genomic DNA from the plasmid preparation.
As the expected band was present in line 2, we further used this plasmid for transformation. Therefore, we used this plasmid containing the ori R6K and a gene for a kanamycin resistance and transformed it in competent E. coli BW29427 and E. coli DH5α [Sambrook J, Fritsch EF, and Maniatis T (1989), Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.]. Previously we used the same batch of competent E. coli DH5α for transformations to test their transformation capability.
The shown transformation in fig. 2 was made with the same protocol and batch of E. coli Dh5α as a positive control, as it contains ori pMB1 which is known to work in E. coli Dh5α. Transformation in E. coli BW29427 was successful and there was no growth of E. coli DH5α due to the selection of nonresistant cells on LB-Kan agar plates.
Afterwards we made a plasmid preparation of transformed E. coli BW29427 to get quantitative data for plasmid concentrations. Therefore, for a plasmid preparation of four batches of E. coli BW29427 was performed. OD(600) of overnight cultures was adjusted to 2,55 and a commercial kit was used for plasmid preparation (Nucleospin Macherey-Nagel). Final DNA concentrations were measured via nano-drop. The resulting table is shown below:
Table 1: DNA concentrations [ng/μL] after plasmid preparation of transformed E. coli BW29427.
| Culture number | DNA concentration [ng/μL] |
|---|---|
| 1 | 12,4 |
| 2 | 14,6 |
| 3 | 13,9 |
| 4 | 14,5 |
To compare this result, we prepped plasmids from DH5α containing ori pMB1, where we archieved DNA concentrations between 190 – 280 ng/μL from the same volume of culture normed to the same OD(600) of 2,55. Plasmid concentration is low compared to ori pMB1 but expected as the copy number of ori R6K is 15 – 20 and the copy number of ori pMB1 is 500 – 700 [Morgan, K. (2014, February 14). Plasmids 101: Origin of Replication. Retrieved from https://blog.addgene.org/plasmid-101-origin-of-replication], meaning ori R6K is not viable for amplification of plasmids but works if high copy numbers are not needed.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |

 1 Registry Star
1 Registry Star