Conjugation
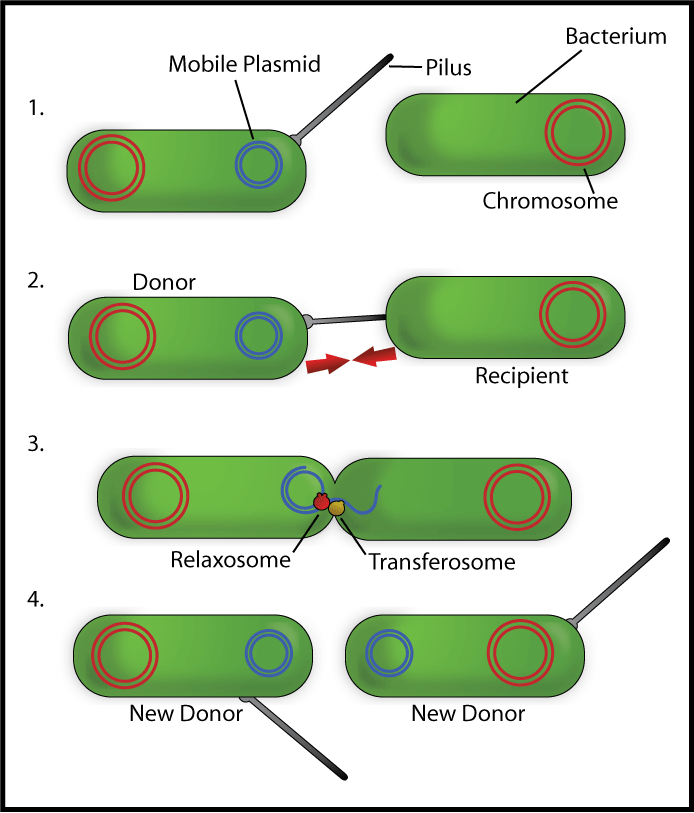
Bacterial conjugation is the transfer of genetic material between two bacterial cell via direct cell-to-cell contact. Although often incorrectly characterized as the bacterial equivalent of mating, in fact it is simply the transfer of genetic material from a donor cell to a recipient cell. Bacterial conjugation is also known as a "type IV secretion system".
This behaviour is characterized by the presence of a specialized plasmid (small, circular piece of transferrable DNA most often found within bacteria) known as a conjugative plasmid. The conjugative plasmid holds a distinct and specialized set of coding regions including the following
- OriT (Origin of Transfer): Unlike other plasmids, the conjugative plasmid has its own origin of transfer nic region. This is a specialized portion of the genome where one of the two strands of the circular plasmid DNA is cut to allow for rolling circle replication of the plasmid containing the OriT region into a recipient cell. During rolling circle replication, the cut end of one strand is inserted into the recipient cell while recipient polymerases construct the complementary strand back into a circular piece of DNA.
- TraJ (Transfer genes): The activation of this gene sets off a cascade of other plasmid genes (and thus corresponding protein expressions) which act in concert to form "mating" characteristics in the host bacterial cell. Such salient characteristics include:
- growth of a pilus--a long whip-like apparatus used to hook the two cells together
- fusion of the outer membranes to allow for transfer of genetic material
- formation of "surface exclusion proteins" which prevents the cell containing the conjugative plasmid from mating with other conjugative plasmids of its type
Two of the most well-studied and characterized types of conjugative plasmids are types "F" (from the F plasmid) and "R". The employed nomenclature for conjugative plasmid genes is the gene name first with the type afterwards. For example, the "TraJr" or "TraJR" gene denotes the "TraJ" gene of the "R" type conjugative plasmid.
Note it was previously believed that transfer of genetic material occurred via the pilus itself; now, most think that the transfer of genetic material occurs via a separate direct channel between the two cells.
| Name | Type | Description | Length |
|---|---|---|---|
| BBa_I714000 | Conjugation | R388 oriT ( transfer origin of E.coli IncW group R388 plasmid) | 402 |
| BBa_I714003 | Conjugation | minimum oriT of plasmid R64 ( origin of transfer, R64 is IncI-1 ) | 83 |
| BBa_I714030 | Conjugation | OriT-F (Origin of Transfer for the F-plasmid nic region) | 355 |
| BBa_I714031 | Conjugation | OriT-R (Origin of Transfer for the R751-plasmid nic region) | 278 |
| BBa_I714032 | Conjugation | OriT-S (Origin of Transfer for the pSC101-plasmid nic region) | 131 |
| BBa_J01000 | Coding | TraJF controls conjugative transfer in F plasmid | 690 |
| BBa_J01001 | Coding | TraJr protein (controls conjugative transfer in RP4 plasmid) | 378 |
| BBa_J01002 | Conjugation | OriT-F (Origin of Transfer for the F-plasmid nic region) | 365 |
| BBa_J01003 | Conjugation | OriT-R (Origin of transfer for the R-plasmid nic region) | 371 |
| BBa_J01077 | Plasmid | F plasmid (x-OriT, x-TraJ) | |
| BBa_J01078 | Plasmid | R plasmid (x-OriT, x-TraJ) | |
| BBa_J107185 | Conjugation | oriT from RK2 | 354 |
| BBa_J23046 | Conjugation | put in later - SIL strain | |
| BBa_J61030 | Composite | [OriTr][R6K] | 785 |
| BBa_K1053123 | Conjugation | Split EYFP N-terminal domain fused with HTLV-1 Rex peptide | 606 |
| BBa_K1053124 | Conjugation | Split EYFP C-terminal domain fused with λN peptide | 297 |
| BBa_K1084999 | Conjugation | GGA_vector | 125 |
| BBa_K109550 | Coding | Bacterial conjugative regulator KorA | 306 |
| BBa_K109551 | Coding | TrbA transcription factor for the Tra2 operon in incP alpha conjugation | 366 |
| BBa_K125320 | Conjugation | OriT-RP4 (Origin of transfer for the RP4 plasmid) | 99 |
| BBa_K125350 | Conjugation | oriTr with lac promoter and lacZ | 979 |
| BBa_K125360 | Conjugation | oriT with lac promoter and lacZ | 707 |
| BBa_K1439000 | Conjugation | Origin of transfer for the RP4-plasmid nic region. | 350 |
| BBa_K150006 | Conjugation | oriT | 471 |
| BBa_K1510119 | Conjugation | A truncated INPNC anchor protein that binds fragments on the cell surface | 1564 |
| BBa_K1510214 | Conjugation | Dispersin generator circuit | 2053 |
| BBa_K1510409 | Conjugation | A Steptococcus Mutants adhesive complex. | 1780 |
| BBa_K1510603 | Conjugation | Dispersin generator ,with constitutive promoter plus yebf | 1916 |
| BBa_K1510624 | Conjugation | Truncated lysostaphin coding sequence plus strong promoter and yebf | 1331 |
| BBa_K1510816 | Conjugation | A Steptococcus Mutants adhesive complex. | 1651 |
| BBa_K1510831 | Conjugation | Fragrance generator part2: alcohol acetyltransferase I and RBS | 1599 |
| BBa_K1510904 | Conjugation | Truncated lysostaphin coding circuit | 1468 |
| BBa_K1510915 | Conjugation | ketoisocaproate decarboxylase (THI3) Circuit | 1985 |
| BBa_K1510922 | Conjugation | Fragrance generator part1: ketoisocaproate decarboxylase (THI3) and RBS | 1848 |
| BBa_K1510925 | Conjugation | Fragrance generator3: branched-chain amino acid transaminase (BAT2) and RBS | 1152 |
| BBa_K1682014 | Conjugation | Conjugation(RP4) plasmid with partial GFP | 1395 |
| BBa_K175000 | Coding | trbC (pilin) of IncP beta R751 plasmid | 468 |
| BBa_K175001 | Coding | trbK (entry exclusion) of IncP beta R751 plasmid | 231 |
| BBa_K188777 | Conjugation | aka | 879 |
| BBa_K188778 | Conjugation | as | 864 |
| BBa_K2144101 | Plasmid_Backbone | Vector containing Hisx6 and Sortase tag LPETGG | 2117 |
| BBa_K2657002 | Conjugation | Rp4 Origin of Transfer PhytoBrick | 131 |
| BBa_K2745002 | Conjugation | linker | 15 |
| BBa_K2759003 | Conjugation | Bacterioferritin (BFR) with sfGFP conjugation | 1203 |
| BBa_K2791011 | Conjugation | Translational coupling device, weak. | 32 |
| BBa_K2791012 | Conjugation | Translational coupling device, strong. | 32 |
| BBa_K2791013 | Conjugation | Translational coupling device, medium. | 32 |
| BBa_K2791014 | Conjugation | Translational coupling device, very weak. | 32 |
| BBa_K2796006 | Conjugation | P2A | 66 |
| BBa_K2796007 | Conjugation | T2A | 63 |
| BBa_K2913002 | Conjugation | A conjugation between Pluc anad HucO | 11 |
| BBa_K2913008 | Conjugation | A referenced conjugation between Pluc and CDS sequence | 35 |
| BBa_K2961003 | Conjugation | gRNA / crRNA fixed-loop repeat for Cpf1 / Cas12a | 21 |
| BBa_K2976010 | Conjugation | Improved T2A | 63 |
| BBa_K2976011 | Conjugation | (Gly4Ser)3 linker | 30 |
| BBa_K2976012 | Conjugation | Gly-Ser-Ser linker | 9 |
| BBa_K3190206 | Conjugation | Linker | 24 |
| BBa_K3819025 | Conjugation | Pro/Thr-rich Linker | 108 |
| BBa_K3875021 | Conjugation | Cas9 binding | 34 |
| BBa_K3946006 | Conjugation | L2NC silica tag+term (CT) | 4461 |
| BBa_K3946007 | Conjugation | L2NC+linker+term (CT) | 4516 |
| BBa_K3946044 | Conjugation | Sb7 silica tag (C)-PETase (W159H/ S238F) | 902 |
| BBa_K3946045 | Conjugation | Sb7 silica tag (C)-LCC-WCCG variant | 811 |
| BBa_K4103001 | Conjugation | pIP501 transfer origin | 378 |
| BBa_K4103016 | Conjugation | 5'lacA | 508 |
| BBa_K4122007 | Conjugation | spytag | |
| BBa_K4122008 | Conjugation | Spycatcher | |
| BBa_K4122009 | Conjugation | Snooptag | |
| BBa_K4122010 | Conjugation | Snoopcatcher | |
| BBa_K4170009 | Conjugation | Serine-serine-glycine flexible linker | 9 |
| BBa_K4580004 | Conjugation | NdmB-6x his-GFP | 1806 |
| BBa_K4587106 | Conjugation | GS linker | 48 |
| BBa_K4643003 | Conjugation | incP origin of transfer | 109 |
| BBa_K4651000 | Conjugation | sdjhf | |
| BBa_K4696019 | Conjugation | P2A | 57 |
| BBa_K4761030 | Conjugation | linker1 | 15 |
| BBa_K4761031 | Conjugation | linker2 | 45 |
| BBa_K4761032 | Conjugation | linker3 | 24 |
| BBa_K4761033 | Conjugation | linker4 | 15 |
| BBa_K4761034 | Conjugation | linker5 | 45 |
| BBa_K4761035 | Conjugation | linker6 | 15 |
| BBa_K4765023 | RNA | PmeI linker | 27 |
| BBa_K4971011 | Conjugation | Linker protein sequence with starting codon | 30 |
| BBa_K5096400 | Conjugation | pCas9 | 8437 |
| BBa_K5170000 | Conjugation | A novel PROTAC using BBa_K5170004 against 231pTau | 66 |
| BBa_K5170001 | Conjugation | A novel PROTAC using BBa_K5170002 against 231pTau | 66 |
| BBa_K5170005 | Conjugation | A novel PROTAC using BBa_K5170003 against 231pTau | 66 |
| BBa_K543016 | Conjugation | Bacteriophage Lysis Cassette S105, R, and Rz under Tet promoter (conjugative) | 1838 |
| BBa_K543020 | Conjugation | OriTr-GFP | 1254 |
| BBa_K543030 | Conjugation | [I714030][J23066][I714030] (Constitutive expression) | 1105 |
| BBa_K543032 | Conjugation | assay system of killing by conjugation | 3615 |
| BBa_K5436020 | Conjugation | BamHI Linker | 6 |
| BBa_K5465999 | Plasmid | ONERING-TetO | 6902 |
| BBa_K640002 | Conjugation | oriT - Interspecies origin of transfer | 260 |
| BBa_K640004 | Conjugation | Device for conjugation of plasmids from E. coli to pseudomonas | 642 |
| BBa_K640005 | Conjugation | Device for conjugating plasmids from E. coli to pseudomonas | 753 |
| BBa_K808028 | Signalling | PhoA: signal sequence for peri plasmic expression | 63 |
| BBa_K873003 | Conjugation | conjugative transfer of RFG | 1302 |
| BBa_M1355 | Conjugation | RS1 | 720 |
| BBa_M1356 | Conjugation | RS2 | 709 |

|

|
Yifan Yang, Mingzhi Qu and the 2007 Peking Yniveristy iGEM team designed the parts BBa_I714000, BBa_I714003, BBa_I714030, BBa_I714031 and BBa_I714032. | The 2005 Berkeley iGEM team designed the parts BBa_J01000-BBa_J01003 and BBa_J01077-BBa_J01078. | ||

|
Chris Anderson and the 2006 Berkeley iGEM team design the parts BBa_J23046. | 
|
Ingrid Swanson and the 2008 University of Washington iGEM team designed the parts BBa_K109550 and BBa_K109551. | 
|
Margaret Ruzicki and the 2008 University of Hawaii iGEM team designed the parts Part:BBa_K125320, Part:BBa_K125350, and Part:BBa_K125360. |
References
<biblio>
- Frost pmid=7915817
- Jaenecke pmid=8858584
- Lawley pmid=12855161
- Lessl pmid=8407818
- Miller pmid=2993231
- Martinez-Morales pmid=10559184
- Wilkins pmid=12220405
</biblio>
- [http://en.wikipedia.org/wiki/Bacterial_conjugation Wikipedia entry on Bacterial Conjugation]
- 2005 UC Berkeley iGEM team [http://2006.igem.org/Berkeley_2005 project] and [http://2006.igem.org/Berkeley_Protocols protocols]
