Difference between revisions of "Part:BBa E1010"
| Line 254: | Line 254: | ||
<partinfo>BBa_E1010 SequenceAndFeatures</partinfo> | <partinfo>BBa_E1010 SequenceAndFeatures</partinfo> | ||
| − | + | ||
{|width='80%' style='border:1px solid gray' | {|width='80%' style='border:1px solid gray' | ||
|- | |- | ||
|width='10%'| | |width='10%'| | ||
| − | + | |}; | |
===Parts table=== | ===Parts table=== | ||
<html><!--- Please copy this table containing parameters for BBa_E1010 at the end of the parametrs section ahead of the references. ---><style type="text/css">table#AutoAnnotator {border:1px solid black; width:100%; border-collapse:collapse;} th#AutoAnnotatorHeader { border:1px solid black; width:100%; background-color: rgb(221, 221, 221);} td.AutoAnnotator1col { width:100%; border:1px solid black; } span.AutoAnnotatorSequence { font-family:'Courier New', Arial; } td.AutoAnnotatorSeqNum { text-align:right; width:2%; } td.AutoAnnotatorSeqSeq { width:98% } td.AutoAnnotatorSeqFeat1 { width:3% } td.AutoAnnotatorSeqFeat2a { width:27% } td.AutoAnnotatorSeqFeat2b { width:97% } td.AutoAnnotatorSeqFeat3 { width:70% } table.AutoAnnotatorNoBorder { border:0px; width:100%; border-collapse:collapse; } table.AutoAnnotatorWithBorder { border:1px solid black; width:100%; border-collapse:collapse; } td.AutoAnnotatorOuterAmino { border:0px solid black; width:20% } td.AutoAnnotatorInnerAmino { border:1px solid black; width:50% } td.AutoAnnotatorAminoCountingOuter { border:1px solid black; width:40%; } td.AutoAnnotatorBiochemParOuter { border:1px solid black; width:60%; } td.AutoAnnotatorAminoCountingInner1 { width: 7.5% } td.AutoAnnotatorAminoCountingInner2 { width:62.5% } td.AutoAnnotatorAminoCountingInner3 { width:30% } td.AutoAnnotatorBiochemParInner1 { width: 5% } td.AutoAnnotatorBiochemParInner2 { width:55% } td.AutoAnnotatorBiochemParInner3 { width:40% } td.AutoAnnotatorCodonUsage1 { width: 3% } td.AutoAnnotatorCodonUsage2 { width:14.2% } td.AutoAnnotatorCodonUsage3 { width:13.8% } td.AutoAnnotatorAlignment1 { width: 3% } td.AutoAnnotatorAlignment2 { width: 10% } td.AutoAnnotatorAlignment3 { width: 87% } td.AutoAnnotatorLocalizationOuter {border:1px solid black; width:40%} td.AutoAnnotatorGOOuter {border:1px solid black; width:60%} td.AutoAnnotatorLocalization1 { width: 7.5% } td.AutoAnnotatorLocalization2 { width: 22.5% } td.AutoAnnotatorLocalization3 { width: 70% } td.AutoAnnotatorGO1 { width: 5% } td.AutoAnnotatorGO2 { width: 35% } td.AutoAnnotatorGO3 { width: 60% } td.AutoAnnotatorPredFeat1 { width:3% } td.AutoAnnotatorPredFeat2a { width:27% } td.AutoAnnotatorPredFeat3 { width:70% } div.AutoAnnotator_trans { position:absolute; background:rgb(11,140,143); background-color:rgba(11,140,143, 0.8); height:5px; top:100px; } div.AutoAnnotator_sec_helix { position:absolute; background:rgb(102,0,102); background-color:rgba(102,0,102, 0.8); height:5px; top:110px; } div.AutoAnnotator_sec_strand { position:absolute; background:rgb(245,170,26); background-color:rgba(245,170,26, 1); height:5px; top:110px; } div.AutoAnnotator_acc_buried { position:absolute; background:rgb(89,168,15); background-color:rgba(89,168,15, 0.8); height:5px; top:120px; } div.AutoAnnotator_acc_exposed { position:absolute; background:rgb(0, 0, 255); background-color:rgba(0, 0, 255, 0.8); height:5px; top:120px; } div.AutoAnnotator_dis { position:absolute; text-align:center; font-family:Arial,Helvetica,sans-serif; background:rgb(255, 200, 0); background-color:rgba(255, 200, 0, 1); height:16px; width:16px; top:80px; border-radius:50%; } </style><div id='AutoAnnotator_container_1397080705152'><table id="AutoAnnotator"><tr><!-- Time stamp in ms since 1/1/1970 1397080705152 --><th id="AutoAnnotatorHeader" colspan="2">Protein data table for BioBrick <a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_E1010">BBa_E1010</a> automatically created by the <a href="http://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">BioBrick-AutoAnnotator</a> version 1.0</th></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Nucleotide sequence</strong> in <strong>RFC 10</strong>: (underlined part encodes the protein)<br><span class="AutoAnnotatorSequence"> <u>ATGGCTTCC ... ACCGGTGCT</u>TAATAACGCTGATAGTGCTAGTGTAGATCGC</span><br> <strong>ORF</strong> from nucleotide position 1 to 675 (excluding stop-codon)</td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid sequence:</strong> (RFC 25 scars in shown in bold, other sequence features underlined; both given below)<br><span class="AutoAnnotatorSequence"><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqNum">1 <br>101 <br>201 </td><td class="AutoAnnotatorSeqSeq">MASSEDVIKEFMRFKVRMEGSVNGHEFEIEGEGEGRPYEGTQTAKLKVTKGGPLPFAWDILSPQFQYGSKAYVKHPADIPDYLKLSFPEGFKWERVMNFE<br>DGGVVTVTQDSSLQDGEFIYKVKLRGTNFPSDGPVMQKKTMGWEASTERMYPEDGALKGEIKMRLKLKDGGHYDAEVKTTYMAKKPVQLPGAYKTDIKLD<br>ITSHNEDYTIVEQYERAEGRHSTGA*</td></tr></table></span></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Sequence features:</strong> (with their position in the amino acid sequence, see the <a href="http://2013.igem.org/Team:TU-Munich/Results/Software/FeatureList">list of supported features</a>)<table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqFeat1"></td><td class="AutoAnnotatorSeqFeat2b">None of the supported features appeared in the sequence</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid composition:</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Ala (A)</td><td class="AutoAnnotatorInnerAmino">12 (5.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Arg (R)</td><td class="AutoAnnotatorInnerAmino">9 (4.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asn (N)</td><td class="AutoAnnotatorInnerAmino">4 (1.8%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asp (D)</td><td class="AutoAnnotatorInnerAmino">14 (6.2%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Cys (C)</td><td class="AutoAnnotatorInnerAmino">0 (0.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gln (Q)</td><td class="AutoAnnotatorInnerAmino">8 (3.6%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Glu (E)</td><td class="AutoAnnotatorInnerAmino">22 (9.8%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gly (G)</td><td class="AutoAnnotatorInnerAmino">23 (10.2%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">His (H)</td><td class="AutoAnnotatorInnerAmino">5 (2.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ile (I)</td><td class="AutoAnnotatorInnerAmino">9 (4.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Leu (L)</td><td class="AutoAnnotatorInnerAmino">12 (5.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Lys (K)</td><td class="AutoAnnotatorInnerAmino">22 (9.8%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Met (M)</td><td class="AutoAnnotatorInnerAmino">9 (4.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Phe (F)</td><td class="AutoAnnotatorInnerAmino">10 (4.4%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Pro (P)</td><td class="AutoAnnotatorInnerAmino">12 (5.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ser (S)</td><td class="AutoAnnotatorInnerAmino">12 (5.3%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Thr (T)</td><td class="AutoAnnotatorInnerAmino">14 (6.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Trp (W)</td><td class="AutoAnnotatorInnerAmino">3 (1.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Tyr (Y)</td><td class="AutoAnnotatorInnerAmino">11 (4.9%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Val (V)</td><td class="AutoAnnotatorInnerAmino">14 (6.2%)</td></tr></table></td></tr></table></td></tr><tr><td class="AutoAnnotatorAminoCountingOuter"><strong>Amino acid counting</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Total number:</td><td class="AutoAnnotatorAminoCountingInner3">225</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Positively charged (Arg+Lys):</td><td class="AutoAnnotatorAminoCountingInner3">31 (13.8%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Negatively charged (Asp+Glu):</td><td class="AutoAnnotatorAminoCountingInner3">36 (16.0%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Aromatic (Phe+His+Try+Tyr):</td><td class="AutoAnnotatorAminoCountingInner3">29 (12.9%)</td></tr></table></td><td class="AutoAnnotatorBiochemParOuter"><strong>Biochemical parameters</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Atomic composition:</td><td class="AutoAnnotatorBiochemParInner3">C<sub>1135</sub>H<sub>1749</sub>N<sub>299</sub>O<sub>347</sub>S<sub>9</sub></td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Molecular mass [Da]:</td><td class="AutoAnnotatorBiochemParInner3">25423.7</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Theoretical pI:</td><td class="AutoAnnotatorBiochemParInner3">5.65</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Extinction coefficient at 280 nm [M<sup>-1</sup> cm<sup>-1</sup>]:</td><td class="AutoAnnotatorBiochemParInner3">32890 / 32890 (all Cys red/ox)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges</strong> <input type='button' id='hydrophobicity_charge_button' onclick='show_or_hide_plot_1397080705152()' value='Show'><span id="hydrophobicity_charge_explanation"></span><div id="hydrophobicity_charge_container" style='display:none'><div id="hydrophobicity_charge_placeholder0" style="width:100%;height:150px"></div><div id="hydrophobicity_charge_placeholder1" style="width:100%;height:150px"></div></div></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Codon usage</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Organism:</td><td class="AutoAnnotatorCodonUsage3"><i>E. coli</i></td><td class="AutoAnnotatorCodonUsage3"><i>B. subtilis</i></td><td class="AutoAnnotatorCodonUsage3"><i>S. cerevisiae</i></td><td class="AutoAnnotatorCodonUsage3"><i>A. thaliana</i></td><td class="AutoAnnotatorCodonUsage3"><i>P. patens</i></td><td class="AutoAnnotatorCodonUsage3">Mammals</td></tr><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Codon quality (<a href="http://en.wikipedia.org/wiki/Codon_Adaptation_Index">CAI</a>):</td><td class="AutoAnnotatorCodonUsage3">excellent (0.84)</td><td class="AutoAnnotatorCodonUsage3">good (0.72)</td><td class="AutoAnnotatorCodonUsage3">good (0.68)</td><td class="AutoAnnotatorCodonUsage3">good (0.74)</td><td class="AutoAnnotatorCodonUsage3">good (0.78)</td><td class="AutoAnnotatorCodonUsage3">good (0.71)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Alignments</strong> (obtained from <a href='http://predictprotein.org'>PredictProtein.org</a>)<table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorAlignment1"></td><td class="AutoAnnotatorAlignment2">SwissProt:</td><td class="AutoAnnotatorAlignment3"><a href='http://www.uniprot.org/uniprot/Q9U6Y8'>Q9U6Y8</a> (86% identity on 221 AAs), <a href='http://www.uniprot.org/uniprot/P83690'>P83690</a> (63% identity on 215 AAs)</td></tr><tr><td class="AutoAnnotatorAlignment1"></td><td class="AutoAnnotatorAlignment2">TrEML:</td><td class="AutoAnnotatorAlignment3"><a href='http://www.uniprot.org/uniprot/Q5S3G8'>Q5S3G8</a> (95% identity on 225 AAs), <a href='http://www.uniprot.org/uniprot/D0VWW2'>D0VWW2</a> (94% identity on 220 AAs)</td></tr><tr><td class="AutoAnnotatorAlignment1"></td><td class="AutoAnnotatorAlignment2">PDB:</td><td class="AutoAnnotatorAlignment3"><a href='http://www.rcsb.org/pdb/explore/explore.do?structureId=2h5q'>2h5q</a> (94% identity on 216 AAs), <a href='http://www.rcsb.org/pdb/explore/explore.do?structureId=2qlg'>2qlg</a> (94% identity on 215 AAs)</td></tr></table></td></tr><tr><th id='AutoAnnotatorHeader' colspan="2"><strong>Predictions</strong> (obtained from <a href='http://predictprotein.org'>PredictProtein.org</a>)</th></tr><tr><td class="AutoAnnotatorLocalizationOuter"><strong>Subcellular Localization</strong> (reliability in brackets)<table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorLocalization1"></td><td class="AutoAnnotatorLocalization2">Archaea:</td><td class="AutoAnnotatorLocalization3">secreted (100%)</td></tr><tr><td class="AutoAnnotatorLocalization1"></td><td class="AutoAnnotatorLocalization2">Bacteria:</td><td class="AutoAnnotatorLocalization3">cytosol (52%)</td></tr><tr><td class="AutoAnnotatorLocalization1"></td><td class="AutoAnnotatorLocalization2">Eukarya:</td><td class="AutoAnnotatorLocalization3">cytosol (20%)</td></tr></table></td><td class="AutoAnnotatorGOOuter"><strong>Gene Ontology</strong> (reliability in brackets)<br><table class="AutoAnnotatorNoBorder"><tr><td class='AutoAnnotatorGO1'></td><td class='AutoAnnotatorGO2'>Molecular Function Ontology:</td><td class='AutoAnnotatorGO3'> - </td></tr><tr><td class='AutoAnnotatorGO1'></td><td class='AutoAnnotatorGO2'>Biological Process Ontology:</td><td class='AutoAnnotatorGO3'><a href='http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0018298'>GO:0018298</a> (40%), <a href='http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0008218'>GO:0008218</a> (27%)</td></tr><tr><td class='AutoAnnotatorGO1'> </td><td class='AutoAnnotatorGO2'> </td><td class='AutoAnnotatorGO3'> </td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Predicted features:</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorPredFeat1"></td><td class="AutoAnnotatorPredFeat2a">Disulfid bridges:</td><td class="AutoAnnotatorPredFeat3"> - </td></tr><tr><td class="AutoAnnotatorPredFeat1"></td><td class="AutoAnnotatorPredFeat2a">Transmembrane helices:</td><td class="AutoAnnotatorPredFeat3"> - </td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"> The BioBrick-AutoAnnotator was created by <a href="http://2013.igem.org/Team:TU-Munich">TU-Munich 2013</a> iGEM team. For more information please see the <a href="http://2013.igem.org/Team:TU-Munich/Results/Software">documentation</a>.<br>If you have any questions, comments or suggestions, please leave us a <a href="http://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">comment</a>.</td></tr></table></div><br><!-- IMPORTANT: DON'T REMOVE THIS LINE, OTHERWISE NOT SUPPORTED FOR IE BEFORE 9 --><!--[if lte IE 8]><script language="javascript" type="text/javascript" src="http://2013.igem.org/Team:TU-Munich/excanvas.js"></script><![endif]--><script type='text/javascript' src='http://code.jquery.com/jquery-1.10.0.min.js'></script><script type='text/javascript' src='http://2013.igem.org/Team:TU-Munich/Flot.js?action=raw&ctype=text/js'></script><script>var jqAutoAnnotator = jQuery.noConflict(true);function show_or_hide_plot_1397080705152(){hydrophobicity_datapoints = [[2.5,-0.28],[3.5,-1.36],[4.5,-0.88],[5.5,0.18],[6.5,-0.44],[7.5,-0.44],[8.5,0.82],[9.5,0.36],[10.5,-1.44],[11.5,-0.10],[12.5,-0.18],[13.5,0.10],[14.5,-1.18],[15.5,0.10],[16.5,-1.16],[17.5,-0.46],[18.5,-1.46],[19.5,0.28],[20.5,-0.80],[21.5,-0.18],[22.5,-0.74],[23.5,-1.28],[24.5,-1.56],[25.5,-1.56],[26.5,-0.58],[27.5,-0.64],[28.5,-0.02],[29.5,-1.28],[30.5,-0.66],[31.5,-2.26],[32.5,-1.64],[33.5,-2.46],[34.5,-2.08],[35.5,-2.26],[36.5,-2.26],[37.5,-2.26],[38.5,-1.50],[39.5,-1.88],[40.5,-1.76],[41.5,-0.70],[42.5,-1.40],[43.5,-0.50],[44.5,-0.58],[45.5,0.40],[46.5,-0.10],[47.5,-0.10],[48.5,-0.94],[49.5,-0.24],[50.5,-1.40],[51.5,-0.50],[52.5,-0.04],[53.5,0.60],[54.5,1.04],[55.5,1.18],[56.5,-0.28],[57.5,0.94],[58.5,1.14],[59.5,0.62],[60.5,0.48],[61.5,0.48],[62.5,0.14],[63.5,-1.32],[64.5,-1.42],[65.5,-1.18],[66.5,-0.64],[67.5,-1.98],[68.5,-0.92],[69.5,-0.92],[70.5,-0.00],[71.5,-0.62],[72.5,-0.48],[73.5,-1.16],[74.5,-0.54],[75.5,-2.08],[76.5,-0.40],[77.5,-0.08],[78.5,-0.46],[79.5,-1.08],[80.5,0.38],[81.5,-1.30],[82.5,-0.22],[83.5,0.32],[84.5,1.14],[85.5,0.06],[86.5,0.14],[87.5,-0.70],[88.5,0.02],[89.5,-1.32],[90.5,-1.18],[91.5,-1.18],[92.5,-2.00],[93.5,-1.72],[94.5,-0.56],[95.5,-1.08],[96.5,0.18],[97.5,0.38],[98.5,-1.16],[99.5,-1.62],[100.5,-1.00],[101.5,-0.72],[102.5,0.82],[103.5,1.38],[104.5,2.30],[105.5,2.24],[106.5,0.70],[107.5,-0.84],[108.5,-0.86],[109.5,-1.86],[110.5,-0.96],[111.5,-0.96],[112.5,-0.96],[113.5,-0.88],[114.5,-1.42],[115.5,-1.62],[116.5,-0.02],[117.5,0.42],[118.5,-0.28],[119.5,1.26],[120.5,-0.08],[121.5,-0.22],[122.5,-0.86],[123.5,-0.16],[124.5,-1.14],[125.5,-1.06],[126.5,-1.26],[127.5,-0.68],[128.5,-0.76],[129.5,-1.32],[130.5,-0.70],[131.5,-1.58],[132.5,-0.42],[133.5,0.12],[134.5,0.12],[135.5,-0.58],[136.5,-1.04],[137.5,-2.02],[138.5,-2.02],[139.5,-1.40],[140.5,-0.80],[141.5,-0.72],[142.5,-0.22],[143.5,-0.76],[144.5,-0.82],[145.5,-1.34],[146.5,-1.54],[147.5,-1.52],[148.5,-1.62],[149.5,-1.80],[150.5,-1.80],[151.5,-1.60],[152.5,-2.06],[153.5,-1.44],[154.5,-0.36],[155.5,-0.44],[156.5,0.18],[157.5,-0.44],[158.5,0.10],[159.5,-1.44],[160.5,-0.28],[161.5,-1.10],[162.5,0.36],[163.5,-1.32],[164.5,0.22],[165.5,-0.94],[166.5,-0.74],[167.5,-1.58],[168.5,-0.88],[169.5,-2.28],[170.5,-1.76],[171.5,-1.76],[172.5,-1.32],[173.5,-1.94],[174.5,-0.46],[175.5,-0.98],[176.5,-0.42],[177.5,-0.92],[178.5,-0.48],[179.5,-0.94],[180.5,0.20],[181.5,-0.44],[182.5,-1.08],[183.5,-1.14],[184.5,-0.68],[185.5,-1.74],[186.5,-0.20],[187.5,0.26],[188.5,0.50],[189.5,0.02],[190.5,0.46],[191.5,-1.08],[192.5,-0.90],[193.5,-1.52],[194.5,-0.98],[195.5,-1.50],[196.5,0.04],[197.5,-0.52],[198.5,1.08],[199.5,0.04],[200.5,0.66],[201.5,-0.74],[202.5,-0.74],[203.5,-2.34],[204.5,-2.90],[205.5,-3.00],[206.5,-2.50],[207.5,-0.90],[208.5,0.64],[209.5,0.64],[210.5,0.20],[211.5,0.08],[212.5,-1.52],[213.5,-3.26],[214.5,-2.20],[215.5,-2.20],[216.5,-2.02],[217.5,-2.22],[218.5,-1.96],[219.5,-2.48],[220.5,-1.92],[221.5,-1.92],[222.5,-0.66]];charge_datapoints = [[2.5,-0.20],[3.5,-0.40],[4.5,-0.40],[5.5,-0.40],[6.5,-0.20],[7.5,-0.20],[8.5,0.00],[9.5,0.00],[10.5,0.20],[11.5,0.00],[12.5,0.40],[13.5,0.40],[14.5,0.60],[15.5,0.40],[16.5,0.20],[17.5,0.00],[18.5,0.00],[19.5,-0.20],[20.5,-0.20],[21.5,0.00],[22.5,0.10],[23.5,-0.10],[24.5,-0.10],[25.5,-0.30],[26.5,-0.30],[27.5,-0.60],[28.5,-0.40],[29.5,-0.60],[30.5,-0.40],[31.5,-0.60],[32.5,-0.40],[33.5,-0.20],[34.5,0.00],[35.5,0.00],[36.5,0.00],[37.5,0.00],[38.5,-0.20],[39.5,-0.20],[40.5,-0.20],[41.5,0.00],[42.5,0.20],[43.5,0.20],[44.5,0.40],[45.5,0.40],[46.5,0.40],[47.5,0.40],[48.5,0.40],[49.5,0.20],[50.5,0.20],[51.5,0.20],[52.5,0.00],[53.5,0.00],[54.5,0.00],[55.5,0.00],[56.5,-0.20],[57.5,-0.20],[58.5,-0.20],[59.5,-0.20],[60.5,-0.20],[61.5,0.00],[62.5,0.00],[63.5,0.00],[64.5,0.00],[65.5,0.00],[66.5,0.00],[67.5,0.20],[68.5,0.20],[69.5,0.20],[70.5,0.20],[71.5,0.40],[72.5,0.30],[73.5,0.30],[74.5,0.30],[75.5,0.10],[76.5,-0.10],[77.5,-0.20],[78.5,-0.40],[79.5,-0.40],[80.5,-0.20],[81.5,0.00],[82.5,0.00],[83.5,0.20],[84.5,0.20],[85.5,0.20],[86.5,-0.20],[87.5,-0.20],[88.5,-0.20],[89.5,0.00],[90.5,0.00],[91.5,0.00],[92.5,0.20],[93.5,0.20],[94.5,0.00],[95.5,0.00],[96.5,0.20],[97.5,-0.20],[98.5,-0.40],[99.5,-0.40],[100.5,-0.40],[101.5,-0.40],[102.5,-0.20],[103.5,0.00],[104.5,0.00],[105.5,0.00],[106.5,0.00],[107.5,-0.20],[108.5,-0.20],[109.5,-0.20],[110.5,-0.20],[111.5,-0.20],[112.5,-0.20],[113.5,-0.20],[114.5,-0.40],[115.5,-0.40],[116.5,-0.40],[117.5,-0.20],[118.5,0.00],[119.5,0.20],[120.5,0.40],[121.5,0.40],[122.5,0.60],[123.5,0.40],[124.5,0.40],[125.5,0.20],[126.5,0.20],[127.5,0.00],[128.5,0.00],[129.5,-0.20],[130.5,-0.20],[131.5,-0.20],[132.5,-0.20],[133.5,-0.20],[134.5,0.00],[135.5,0.20],[136.5,0.40],[137.5,0.40],[138.5,0.40],[139.5,0.40],[140.5,0.20],[141.5,-0.20],[142.5,-0.20],[143.5,-0.20],[144.5,-0.20],[145.5,-0.40],[146.5,0.00],[147.5,0.00],[148.5,0.00],[149.5,0.00],[150.5,0.00],[151.5,-0.40],[152.5,-0.40],[153.5,-0.40],[154.5,-0.40],[155.5,0.00],[156.5,0.20],[157.5,0.00],[158.5,0.00],[159.5,0.20],[160.5,0.00],[161.5,0.20],[162.5,0.40],[163.5,0.60],[164.5,0.40],[165.5,0.60],[166.5,0.20],[167.5,0.20],[168.5,0.00],[169.5,0.10],[170.5,-0.10],[171.5,-0.10],[172.5,-0.10],[173.5,-0.30],[174.5,-0.40],[175.5,-0.20],[176.5,0.00],[177.5,0.00],[178.5,0.20],[179.5,0.20],[180.5,0.00],[181.5,0.20],[182.5,0.40],[183.5,0.40],[184.5,0.40],[185.5,0.40],[186.5,0.20],[187.5,0.00],[188.5,0.00],[189.5,0.00],[190.5,0.00],[191.5,0.20],[192.5,0.20],[193.5,0.00],[194.5,0.00],[195.5,0.20],[196.5,0.00],[197.5,-0.20],[198.5,0.00],[199.5,0.00],[200.5,-0.20],[201.5,-0.10],[202.5,0.10],[203.5,-0.10],[204.5,-0.30],[205.5,-0.30],[206.5,-0.40],[207.5,-0.40],[208.5,-0.20],[209.5,-0.20],[210.5,-0.20],[211.5,-0.20],[212.5,-0.40],[213.5,-0.20],[214.5,0.00],[215.5,-0.20],[216.5,-0.20],[217.5,0.20],[218.5,0.10],[219.5,0.10],[220.5,0.30],[221.5,0.30],[222.5,0.10]];dis_datapoints = [];trans_datapoints = [];sec_helix_datapoints = [[8,10],[57,66],[79,85]];sec_strand_datapoints = [[13,22],[25,31],[41,49],[93,100],[104,113],[117,126],[148,152],[156,168],[172,183],[195,200],[201,203],[210,221]];acc_exposed_datapoints = [[1,6],[9,10],[13,13],[15,15],[19,19],[23,23],[26,26],[32,32],[34,35],[37,37],[39,39],[41,41],[50,51],[53,53],[70,70],[74,74],[77,78],[81,81],[84,84],[88,90],[92,92],[110,110],[114,117],[128,128],[131,132],[134,134],[137,140],[142,142],[144,145],[148,148],[154,155],[158,158],[168,169],[171,171],[178,178],[182,182],[184,186],[188,188],[191,191],[200,200],[203,203],[205,207],[209,209],[212,212],[215,216],[218,218],[223,225]];acc_buried_datapoints = [[7,8],[12,12],[14,14],[16,16],[18,18],[20,20],[22,22],[24,24],[27,27],[29,29],[31,31],[33,33],[40,40],[44,44],[46,46],[48,48],[54,58],[60,65],[67,68],[71,72],[79,79],[82,83],[87,87],[91,91],[93,93],[95,97],[99,99],[103,107],[111,111],[113,113],[118,118],[120,120],[122,122],[124,124],[126,126],[129,129],[133,133],[135,136],[146,146],[150,151],[156,157],[159,159],[161,161],[163,163],[165,165],[167,167],[173,173],[175,175],[177,177],[179,179],[183,183],[187,187],[189,190],[194,195],[197,197],[199,199],[211,211],[217,217],[219,219]];flot_plot_options = []; flot_plot_options[0] = {grid: {borderWidth: {top: 0,right: 0,bottom: 0,left: 0}},legend: {show: false},xaxes: [{show: true,min: 0,max: 200,ticks: [[0.5, '1'], [24.5, '25'], [49.5, '50'], [74.5, '75'], [99.5, '100'], [124.5, '125'], [149.5, '150'], [174.5, '175'], [199.5, '200']],tickLength: -5}],yaxes: [{show: true,ticks: [[0, '0'], [4.5,'hydro-<br>phobic '], [-4.5,'hydro-<br>philic ']],min: -4.5,max: +4.5,font: {size: 12,lineHeight: 14,style: 'italic',weight: 'bold',family: 'sans-serif',variant: 'small-caps',color: 'rgba(100,149,237,1)'}},{show: true,ticks: [[0, ''], [1,'positive<br> charge'], [-1,'negative<br> charge']],position: 'right',min: -1,max: 1,font: {size: 12,lineHeight: 14,style: 'italic',weight: 'bold',family: 'sans-serif',variant: 'small-caps',color: 'rgba(255,99,71,1)'}}]};number_of_plots = 2;for ( plot_num = 1 ; plot_num < number_of_plots ; plot_num ++){flot_plot_options[plot_num] = jqAutoAnnotator.extend(true, {} ,flot_plot_options[0]);flot_plot_options[plot_num].xaxes = [{min: plot_num*200,max: (plot_num + 1)*200,ticks: [ [plot_num*200 + 0.5, (plot_num*200 + 1).toString()], [plot_num*200 + 24.5, (plot_num*200 + 25).toString()], [plot_num*200 + 49.5, (plot_num*200 + 50).toString()], [plot_num*200 + 74.5, (plot_num*200 + 75).toString()], [plot_num*200 + 99.5, (plot_num*200 + 100).toString()], [plot_num*200 + 124.5, (plot_num*200 + 125).toString()], [plot_num*200 + 149.5, (plot_num*200 + 150).toString()], [plot_num*200 + 174.5, (plot_num*200 + 175).toString()], [plot_num*200 + 199.5, (plot_num*200 + 200).toString()] ],tickLength: -5}];};try {if( jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_button').val() =='Show' ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_container').css('display','block');jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_button').val('Hide');var description_html = '<div id=\'AutoAnnotator_plot_selectors\'>';description_html = description_html + '<br> <input type=\'checkbox\' id=\'hydrophobicity_checkbox\' checked=\'checked\'> Moving average over 5 amino acids for hydrophobicity (<img src=\'https://static.igem.org/mediawiki/2013/e/e9/TUM13_hydrophobicity_icon.png\' alt=\'blue graph\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'charge_checkbox\' checked=\'checked\'> Moving average over 5 amino acids for charge (<img src=\'https://static.igem.org/mediawiki/2013/3/3e/TUM13_charge_icon.png\' alt=\'red graph\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'dis_checkbox\' checked=\'checked\'> Predicted disulfid bridges (<img src=\'https://static.igem.org/mediawiki/2013/2/28/TUM13_dis_icon.png\' alt=\'yellow circle\' height=\'10\'></img>) with the number of the bridge in the center';description_html = description_html + '<br> <input type=\'checkbox\' id=\'trans_checkbox\' checked=\'checked\'> Predicted transmembrane helices (<img src=\'https://static.igem.org/mediawiki/2013/7/78/TUM13_trans_icon.png\' alt=\'turquois bars\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'sec_checkbox\' checked=\'checked\'> Predicted secondary structure: Helices (<img src=\'https://static.igem.org/mediawiki/2013/b/bf/TUM13_helix_icon.png\' alt=\'violet bars\' height=\'10\'></img>) and beta-strands (<img src=\'https://static.igem.org/mediawiki/2013/b/bf/TUM13_strand_icon.png\' alt=\'yellow bars\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'acc_checkbox\' checked=\'checked\'> Predicted solvent accessability: Exposed (<img src=\'https://static.igem.org/mediawiki/2013/1/16/TUM13_exposed_icon.png\' alt=\'blue bars\' height=\'10\'></img>) and buried (<img src=\'https://static.igem.org/mediawiki/2013/0/0b/TUM13_buried_icon.png\' alt=\'green bars\' height=\'10\'></img>) residues';description_html = description_html + '<br></div>';jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_explanation').html(description_html);plot_according_to_selectors_1397080705152();jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #AutoAnnotator_plot_selectors').find('input').click(plot_according_to_selectors_1397080705152);}else{jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_container').css('display','none');jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_button').val('Show');jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_explanation').html('');}}catch(err){txt='There was an error with the button controlling the visibility of the plot.\n';txt=txt+'The originating error is:\n' + err + '\n\n';alert(txt);}};function plot_according_to_selectors_1397080705152(){try{var plot_datasets = [[],[]];if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_checkbox').prop('checked') == true){plot_datasets[0] = { color: 'rgba(100,149,237,1)',data: hydrophobicity_datapoints,label: 'Hydrophobicity',lines: { show: true, fill: true, fillColor: 'rgba(100,149,237,0.1)' },yaxis: 1};}if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #charge_checkbox').prop('checked') == true){plot_datasets[1] = {color: 'rgba(255,99,71,1)',data: charge_datapoints,label: 'Charge',lines: { show: true, fill: true, fillColor: 'rgba(255,99,71,0.1)' },yaxis: 2};}for (plot_num = 0 ; plot_num < number_of_plots ; plot_num ++){jqAutoAnnotator.plot('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder'+ plot_num.toString(), plot_datasets, flot_plot_options[plot_num] );}var screen_width = jqAutoAnnotator('canvas.flot-base').width(); var pos_of_first_tick = 46;var pos_of_last_tick = screen_width - 51;var tick_diff = (screen_width - 97)/199;if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #dis_checkbox').prop('checked') == true){for ( j = 0 ; j < dis_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((dis_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_dis\' style=\'left:' + ((pos_of_first_tick - 8 + (dis_datapoints[j][0] - 1)*tick_diff - Math.floor((dis_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px;\'><b>' + (j+1) + '</b></div>');jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((dis_datapoints[j][1] - 1)/200) ).append('<div class=\'AutoAnnotator_dis\' style=\'left:' + ((pos_of_first_tick - 8 + (dis_datapoints[j][1] - 1)*tick_diff - Math.floor((dis_datapoints[j][1] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px;\'><b>' + (j+1) + '</b></div>');}}if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #trans_checkbox').prop('checked') == true){for ( j = 0 ; j < trans_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((trans_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_trans\' style=\'width:' + (((trans_datapoints[j][1] - trans_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px ;left:' + ((pos_of_first_tick + (trans_datapoints[j][0] - 1.5)*tick_diff - Math.floor((trans_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #sec_checkbox').prop('checked') == true){for ( j = 0 ; j < sec_helix_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((sec_helix_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_sec_helix\' style=\'width:' + (((sec_helix_datapoints[j][1] - sec_helix_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (sec_helix_datapoints[j][0] - 1.5)*tick_diff - Math.floor((sec_helix_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}for ( j = 0 ; j < sec_strand_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((sec_strand_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_sec_strand\' style=\'width:' + (((sec_strand_datapoints[j][1] - sec_strand_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (sec_strand_datapoints[j][0] - 1.5)*tick_diff - Math.floor((sec_strand_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #acc_checkbox').prop('checked') == true){for ( j = 0 ; j < acc_buried_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((acc_buried_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_acc_buried\' style=\'width:' + (((acc_buried_datapoints[j][1] - acc_buried_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (acc_buried_datapoints[j][0] - 1.5)*tick_diff - Math.floor((acc_buried_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}for ( j = 0 ; j < acc_exposed_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((acc_exposed_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_acc_exposed\' style=\'width:' + (((acc_exposed_datapoints[j][1] - acc_exposed_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (acc_exposed_datapoints[j][0] - 1.5)*tick_diff - Math.floor((acc_exposed_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}}catch(err){txt='There was an error while drawing the selected elements for the plot.\n';txt=txt+'The originating error is:\n' + err + '\n\n';throw(txt);}}</script></html> | <html><!--- Please copy this table containing parameters for BBa_E1010 at the end of the parametrs section ahead of the references. ---><style type="text/css">table#AutoAnnotator {border:1px solid black; width:100%; border-collapse:collapse;} th#AutoAnnotatorHeader { border:1px solid black; width:100%; background-color: rgb(221, 221, 221);} td.AutoAnnotator1col { width:100%; border:1px solid black; } span.AutoAnnotatorSequence { font-family:'Courier New', Arial; } td.AutoAnnotatorSeqNum { text-align:right; width:2%; } td.AutoAnnotatorSeqSeq { width:98% } td.AutoAnnotatorSeqFeat1 { width:3% } td.AutoAnnotatorSeqFeat2a { width:27% } td.AutoAnnotatorSeqFeat2b { width:97% } td.AutoAnnotatorSeqFeat3 { width:70% } table.AutoAnnotatorNoBorder { border:0px; width:100%; border-collapse:collapse; } table.AutoAnnotatorWithBorder { border:1px solid black; width:100%; border-collapse:collapse; } td.AutoAnnotatorOuterAmino { border:0px solid black; width:20% } td.AutoAnnotatorInnerAmino { border:1px solid black; width:50% } td.AutoAnnotatorAminoCountingOuter { border:1px solid black; width:40%; } td.AutoAnnotatorBiochemParOuter { border:1px solid black; width:60%; } td.AutoAnnotatorAminoCountingInner1 { width: 7.5% } td.AutoAnnotatorAminoCountingInner2 { width:62.5% } td.AutoAnnotatorAminoCountingInner3 { width:30% } td.AutoAnnotatorBiochemParInner1 { width: 5% } td.AutoAnnotatorBiochemParInner2 { width:55% } td.AutoAnnotatorBiochemParInner3 { width:40% } td.AutoAnnotatorCodonUsage1 { width: 3% } td.AutoAnnotatorCodonUsage2 { width:14.2% } td.AutoAnnotatorCodonUsage3 { width:13.8% } td.AutoAnnotatorAlignment1 { width: 3% } td.AutoAnnotatorAlignment2 { width: 10% } td.AutoAnnotatorAlignment3 { width: 87% } td.AutoAnnotatorLocalizationOuter {border:1px solid black; width:40%} td.AutoAnnotatorGOOuter {border:1px solid black; width:60%} td.AutoAnnotatorLocalization1 { width: 7.5% } td.AutoAnnotatorLocalization2 { width: 22.5% } td.AutoAnnotatorLocalization3 { width: 70% } td.AutoAnnotatorGO1 { width: 5% } td.AutoAnnotatorGO2 { width: 35% } td.AutoAnnotatorGO3 { width: 60% } td.AutoAnnotatorPredFeat1 { width:3% } td.AutoAnnotatorPredFeat2a { width:27% } td.AutoAnnotatorPredFeat3 { width:70% } div.AutoAnnotator_trans { position:absolute; background:rgb(11,140,143); background-color:rgba(11,140,143, 0.8); height:5px; top:100px; } div.AutoAnnotator_sec_helix { position:absolute; background:rgb(102,0,102); background-color:rgba(102,0,102, 0.8); height:5px; top:110px; } div.AutoAnnotator_sec_strand { position:absolute; background:rgb(245,170,26); background-color:rgba(245,170,26, 1); height:5px; top:110px; } div.AutoAnnotator_acc_buried { position:absolute; background:rgb(89,168,15); background-color:rgba(89,168,15, 0.8); height:5px; top:120px; } div.AutoAnnotator_acc_exposed { position:absolute; background:rgb(0, 0, 255); background-color:rgba(0, 0, 255, 0.8); height:5px; top:120px; } div.AutoAnnotator_dis { position:absolute; text-align:center; font-family:Arial,Helvetica,sans-serif; background:rgb(255, 200, 0); background-color:rgba(255, 200, 0, 1); height:16px; width:16px; top:80px; border-radius:50%; } </style><div id='AutoAnnotator_container_1397080705152'><table id="AutoAnnotator"><tr><!-- Time stamp in ms since 1/1/1970 1397080705152 --><th id="AutoAnnotatorHeader" colspan="2">Protein data table for BioBrick <a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_E1010">BBa_E1010</a> automatically created by the <a href="http://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">BioBrick-AutoAnnotator</a> version 1.0</th></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Nucleotide sequence</strong> in <strong>RFC 10</strong>: (underlined part encodes the protein)<br><span class="AutoAnnotatorSequence"> <u>ATGGCTTCC ... ACCGGTGCT</u>TAATAACGCTGATAGTGCTAGTGTAGATCGC</span><br> <strong>ORF</strong> from nucleotide position 1 to 675 (excluding stop-codon)</td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid sequence:</strong> (RFC 25 scars in shown in bold, other sequence features underlined; both given below)<br><span class="AutoAnnotatorSequence"><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqNum">1 <br>101 <br>201 </td><td class="AutoAnnotatorSeqSeq">MASSEDVIKEFMRFKVRMEGSVNGHEFEIEGEGEGRPYEGTQTAKLKVTKGGPLPFAWDILSPQFQYGSKAYVKHPADIPDYLKLSFPEGFKWERVMNFE<br>DGGVVTVTQDSSLQDGEFIYKVKLRGTNFPSDGPVMQKKTMGWEASTERMYPEDGALKGEIKMRLKLKDGGHYDAEVKTTYMAKKPVQLPGAYKTDIKLD<br>ITSHNEDYTIVEQYERAEGRHSTGA*</td></tr></table></span></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Sequence features:</strong> (with their position in the amino acid sequence, see the <a href="http://2013.igem.org/Team:TU-Munich/Results/Software/FeatureList">list of supported features</a>)<table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqFeat1"></td><td class="AutoAnnotatorSeqFeat2b">None of the supported features appeared in the sequence</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid composition:</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Ala (A)</td><td class="AutoAnnotatorInnerAmino">12 (5.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Arg (R)</td><td class="AutoAnnotatorInnerAmino">9 (4.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asn (N)</td><td class="AutoAnnotatorInnerAmino">4 (1.8%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asp (D)</td><td class="AutoAnnotatorInnerAmino">14 (6.2%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Cys (C)</td><td class="AutoAnnotatorInnerAmino">0 (0.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gln (Q)</td><td class="AutoAnnotatorInnerAmino">8 (3.6%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Glu (E)</td><td class="AutoAnnotatorInnerAmino">22 (9.8%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gly (G)</td><td class="AutoAnnotatorInnerAmino">23 (10.2%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">His (H)</td><td class="AutoAnnotatorInnerAmino">5 (2.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ile (I)</td><td class="AutoAnnotatorInnerAmino">9 (4.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Leu (L)</td><td class="AutoAnnotatorInnerAmino">12 (5.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Lys (K)</td><td class="AutoAnnotatorInnerAmino">22 (9.8%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Met (M)</td><td class="AutoAnnotatorInnerAmino">9 (4.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Phe (F)</td><td class="AutoAnnotatorInnerAmino">10 (4.4%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Pro (P)</td><td class="AutoAnnotatorInnerAmino">12 (5.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ser (S)</td><td class="AutoAnnotatorInnerAmino">12 (5.3%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Thr (T)</td><td class="AutoAnnotatorInnerAmino">14 (6.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Trp (W)</td><td class="AutoAnnotatorInnerAmino">3 (1.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Tyr (Y)</td><td class="AutoAnnotatorInnerAmino">11 (4.9%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Val (V)</td><td class="AutoAnnotatorInnerAmino">14 (6.2%)</td></tr></table></td></tr></table></td></tr><tr><td class="AutoAnnotatorAminoCountingOuter"><strong>Amino acid counting</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Total number:</td><td class="AutoAnnotatorAminoCountingInner3">225</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Positively charged (Arg+Lys):</td><td class="AutoAnnotatorAminoCountingInner3">31 (13.8%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Negatively charged (Asp+Glu):</td><td class="AutoAnnotatorAminoCountingInner3">36 (16.0%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Aromatic (Phe+His+Try+Tyr):</td><td class="AutoAnnotatorAminoCountingInner3">29 (12.9%)</td></tr></table></td><td class="AutoAnnotatorBiochemParOuter"><strong>Biochemical parameters</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Atomic composition:</td><td class="AutoAnnotatorBiochemParInner3">C<sub>1135</sub>H<sub>1749</sub>N<sub>299</sub>O<sub>347</sub>S<sub>9</sub></td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Molecular mass [Da]:</td><td class="AutoAnnotatorBiochemParInner3">25423.7</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Theoretical pI:</td><td class="AutoAnnotatorBiochemParInner3">5.65</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Extinction coefficient at 280 nm [M<sup>-1</sup> cm<sup>-1</sup>]:</td><td class="AutoAnnotatorBiochemParInner3">32890 / 32890 (all Cys red/ox)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges</strong> <input type='button' id='hydrophobicity_charge_button' onclick='show_or_hide_plot_1397080705152()' value='Show'><span id="hydrophobicity_charge_explanation"></span><div id="hydrophobicity_charge_container" style='display:none'><div id="hydrophobicity_charge_placeholder0" style="width:100%;height:150px"></div><div id="hydrophobicity_charge_placeholder1" style="width:100%;height:150px"></div></div></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Codon usage</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Organism:</td><td class="AutoAnnotatorCodonUsage3"><i>E. coli</i></td><td class="AutoAnnotatorCodonUsage3"><i>B. subtilis</i></td><td class="AutoAnnotatorCodonUsage3"><i>S. cerevisiae</i></td><td class="AutoAnnotatorCodonUsage3"><i>A. thaliana</i></td><td class="AutoAnnotatorCodonUsage3"><i>P. patens</i></td><td class="AutoAnnotatorCodonUsage3">Mammals</td></tr><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Codon quality (<a href="http://en.wikipedia.org/wiki/Codon_Adaptation_Index">CAI</a>):</td><td class="AutoAnnotatorCodonUsage3">excellent (0.84)</td><td class="AutoAnnotatorCodonUsage3">good (0.72)</td><td class="AutoAnnotatorCodonUsage3">good (0.68)</td><td class="AutoAnnotatorCodonUsage3">good (0.74)</td><td class="AutoAnnotatorCodonUsage3">good (0.78)</td><td class="AutoAnnotatorCodonUsage3">good (0.71)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Alignments</strong> (obtained from <a href='http://predictprotein.org'>PredictProtein.org</a>)<table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorAlignment1"></td><td class="AutoAnnotatorAlignment2">SwissProt:</td><td class="AutoAnnotatorAlignment3"><a href='http://www.uniprot.org/uniprot/Q9U6Y8'>Q9U6Y8</a> (86% identity on 221 AAs), <a href='http://www.uniprot.org/uniprot/P83690'>P83690</a> (63% identity on 215 AAs)</td></tr><tr><td class="AutoAnnotatorAlignment1"></td><td class="AutoAnnotatorAlignment2">TrEML:</td><td class="AutoAnnotatorAlignment3"><a href='http://www.uniprot.org/uniprot/Q5S3G8'>Q5S3G8</a> (95% identity on 225 AAs), <a href='http://www.uniprot.org/uniprot/D0VWW2'>D0VWW2</a> (94% identity on 220 AAs)</td></tr><tr><td class="AutoAnnotatorAlignment1"></td><td class="AutoAnnotatorAlignment2">PDB:</td><td class="AutoAnnotatorAlignment3"><a href='http://www.rcsb.org/pdb/explore/explore.do?structureId=2h5q'>2h5q</a> (94% identity on 216 AAs), <a href='http://www.rcsb.org/pdb/explore/explore.do?structureId=2qlg'>2qlg</a> (94% identity on 215 AAs)</td></tr></table></td></tr><tr><th id='AutoAnnotatorHeader' colspan="2"><strong>Predictions</strong> (obtained from <a href='http://predictprotein.org'>PredictProtein.org</a>)</th></tr><tr><td class="AutoAnnotatorLocalizationOuter"><strong>Subcellular Localization</strong> (reliability in brackets)<table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorLocalization1"></td><td class="AutoAnnotatorLocalization2">Archaea:</td><td class="AutoAnnotatorLocalization3">secreted (100%)</td></tr><tr><td class="AutoAnnotatorLocalization1"></td><td class="AutoAnnotatorLocalization2">Bacteria:</td><td class="AutoAnnotatorLocalization3">cytosol (52%)</td></tr><tr><td class="AutoAnnotatorLocalization1"></td><td class="AutoAnnotatorLocalization2">Eukarya:</td><td class="AutoAnnotatorLocalization3">cytosol (20%)</td></tr></table></td><td class="AutoAnnotatorGOOuter"><strong>Gene Ontology</strong> (reliability in brackets)<br><table class="AutoAnnotatorNoBorder"><tr><td class='AutoAnnotatorGO1'></td><td class='AutoAnnotatorGO2'>Molecular Function Ontology:</td><td class='AutoAnnotatorGO3'> - </td></tr><tr><td class='AutoAnnotatorGO1'></td><td class='AutoAnnotatorGO2'>Biological Process Ontology:</td><td class='AutoAnnotatorGO3'><a href='http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0018298'>GO:0018298</a> (40%), <a href='http://amigo.geneontology.org/cgi-bin/amigo/term_details?term=GO:0008218'>GO:0008218</a> (27%)</td></tr><tr><td class='AutoAnnotatorGO1'> </td><td class='AutoAnnotatorGO2'> </td><td class='AutoAnnotatorGO3'> </td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Predicted features:</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorPredFeat1"></td><td class="AutoAnnotatorPredFeat2a">Disulfid bridges:</td><td class="AutoAnnotatorPredFeat3"> - </td></tr><tr><td class="AutoAnnotatorPredFeat1"></td><td class="AutoAnnotatorPredFeat2a">Transmembrane helices:</td><td class="AutoAnnotatorPredFeat3"> - </td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"> The BioBrick-AutoAnnotator was created by <a href="http://2013.igem.org/Team:TU-Munich">TU-Munich 2013</a> iGEM team. For more information please see the <a href="http://2013.igem.org/Team:TU-Munich/Results/Software">documentation</a>.<br>If you have any questions, comments or suggestions, please leave us a <a href="http://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">comment</a>.</td></tr></table></div><br><!-- IMPORTANT: DON'T REMOVE THIS LINE, OTHERWISE NOT SUPPORTED FOR IE BEFORE 9 --><!--[if lte IE 8]><script language="javascript" type="text/javascript" src="http://2013.igem.org/Team:TU-Munich/excanvas.js"></script><![endif]--><script type='text/javascript' src='http://code.jquery.com/jquery-1.10.0.min.js'></script><script type='text/javascript' src='http://2013.igem.org/Team:TU-Munich/Flot.js?action=raw&ctype=text/js'></script><script>var jqAutoAnnotator = jQuery.noConflict(true);function show_or_hide_plot_1397080705152(){hydrophobicity_datapoints = [[2.5,-0.28],[3.5,-1.36],[4.5,-0.88],[5.5,0.18],[6.5,-0.44],[7.5,-0.44],[8.5,0.82],[9.5,0.36],[10.5,-1.44],[11.5,-0.10],[12.5,-0.18],[13.5,0.10],[14.5,-1.18],[15.5,0.10],[16.5,-1.16],[17.5,-0.46],[18.5,-1.46],[19.5,0.28],[20.5,-0.80],[21.5,-0.18],[22.5,-0.74],[23.5,-1.28],[24.5,-1.56],[25.5,-1.56],[26.5,-0.58],[27.5,-0.64],[28.5,-0.02],[29.5,-1.28],[30.5,-0.66],[31.5,-2.26],[32.5,-1.64],[33.5,-2.46],[34.5,-2.08],[35.5,-2.26],[36.5,-2.26],[37.5,-2.26],[38.5,-1.50],[39.5,-1.88],[40.5,-1.76],[41.5,-0.70],[42.5,-1.40],[43.5,-0.50],[44.5,-0.58],[45.5,0.40],[46.5,-0.10],[47.5,-0.10],[48.5,-0.94],[49.5,-0.24],[50.5,-1.40],[51.5,-0.50],[52.5,-0.04],[53.5,0.60],[54.5,1.04],[55.5,1.18],[56.5,-0.28],[57.5,0.94],[58.5,1.14],[59.5,0.62],[60.5,0.48],[61.5,0.48],[62.5,0.14],[63.5,-1.32],[64.5,-1.42],[65.5,-1.18],[66.5,-0.64],[67.5,-1.98],[68.5,-0.92],[69.5,-0.92],[70.5,-0.00],[71.5,-0.62],[72.5,-0.48],[73.5,-1.16],[74.5,-0.54],[75.5,-2.08],[76.5,-0.40],[77.5,-0.08],[78.5,-0.46],[79.5,-1.08],[80.5,0.38],[81.5,-1.30],[82.5,-0.22],[83.5,0.32],[84.5,1.14],[85.5,0.06],[86.5,0.14],[87.5,-0.70],[88.5,0.02],[89.5,-1.32],[90.5,-1.18],[91.5,-1.18],[92.5,-2.00],[93.5,-1.72],[94.5,-0.56],[95.5,-1.08],[96.5,0.18],[97.5,0.38],[98.5,-1.16],[99.5,-1.62],[100.5,-1.00],[101.5,-0.72],[102.5,0.82],[103.5,1.38],[104.5,2.30],[105.5,2.24],[106.5,0.70],[107.5,-0.84],[108.5,-0.86],[109.5,-1.86],[110.5,-0.96],[111.5,-0.96],[112.5,-0.96],[113.5,-0.88],[114.5,-1.42],[115.5,-1.62],[116.5,-0.02],[117.5,0.42],[118.5,-0.28],[119.5,1.26],[120.5,-0.08],[121.5,-0.22],[122.5,-0.86],[123.5,-0.16],[124.5,-1.14],[125.5,-1.06],[126.5,-1.26],[127.5,-0.68],[128.5,-0.76],[129.5,-1.32],[130.5,-0.70],[131.5,-1.58],[132.5,-0.42],[133.5,0.12],[134.5,0.12],[135.5,-0.58],[136.5,-1.04],[137.5,-2.02],[138.5,-2.02],[139.5,-1.40],[140.5,-0.80],[141.5,-0.72],[142.5,-0.22],[143.5,-0.76],[144.5,-0.82],[145.5,-1.34],[146.5,-1.54],[147.5,-1.52],[148.5,-1.62],[149.5,-1.80],[150.5,-1.80],[151.5,-1.60],[152.5,-2.06],[153.5,-1.44],[154.5,-0.36],[155.5,-0.44],[156.5,0.18],[157.5,-0.44],[158.5,0.10],[159.5,-1.44],[160.5,-0.28],[161.5,-1.10],[162.5,0.36],[163.5,-1.32],[164.5,0.22],[165.5,-0.94],[166.5,-0.74],[167.5,-1.58],[168.5,-0.88],[169.5,-2.28],[170.5,-1.76],[171.5,-1.76],[172.5,-1.32],[173.5,-1.94],[174.5,-0.46],[175.5,-0.98],[176.5,-0.42],[177.5,-0.92],[178.5,-0.48],[179.5,-0.94],[180.5,0.20],[181.5,-0.44],[182.5,-1.08],[183.5,-1.14],[184.5,-0.68],[185.5,-1.74],[186.5,-0.20],[187.5,0.26],[188.5,0.50],[189.5,0.02],[190.5,0.46],[191.5,-1.08],[192.5,-0.90],[193.5,-1.52],[194.5,-0.98],[195.5,-1.50],[196.5,0.04],[197.5,-0.52],[198.5,1.08],[199.5,0.04],[200.5,0.66],[201.5,-0.74],[202.5,-0.74],[203.5,-2.34],[204.5,-2.90],[205.5,-3.00],[206.5,-2.50],[207.5,-0.90],[208.5,0.64],[209.5,0.64],[210.5,0.20],[211.5,0.08],[212.5,-1.52],[213.5,-3.26],[214.5,-2.20],[215.5,-2.20],[216.5,-2.02],[217.5,-2.22],[218.5,-1.96],[219.5,-2.48],[220.5,-1.92],[221.5,-1.92],[222.5,-0.66]];charge_datapoints = [[2.5,-0.20],[3.5,-0.40],[4.5,-0.40],[5.5,-0.40],[6.5,-0.20],[7.5,-0.20],[8.5,0.00],[9.5,0.00],[10.5,0.20],[11.5,0.00],[12.5,0.40],[13.5,0.40],[14.5,0.60],[15.5,0.40],[16.5,0.20],[17.5,0.00],[18.5,0.00],[19.5,-0.20],[20.5,-0.20],[21.5,0.00],[22.5,0.10],[23.5,-0.10],[24.5,-0.10],[25.5,-0.30],[26.5,-0.30],[27.5,-0.60],[28.5,-0.40],[29.5,-0.60],[30.5,-0.40],[31.5,-0.60],[32.5,-0.40],[33.5,-0.20],[34.5,0.00],[35.5,0.00],[36.5,0.00],[37.5,0.00],[38.5,-0.20],[39.5,-0.20],[40.5,-0.20],[41.5,0.00],[42.5,0.20],[43.5,0.20],[44.5,0.40],[45.5,0.40],[46.5,0.40],[47.5,0.40],[48.5,0.40],[49.5,0.20],[50.5,0.20],[51.5,0.20],[52.5,0.00],[53.5,0.00],[54.5,0.00],[55.5,0.00],[56.5,-0.20],[57.5,-0.20],[58.5,-0.20],[59.5,-0.20],[60.5,-0.20],[61.5,0.00],[62.5,0.00],[63.5,0.00],[64.5,0.00],[65.5,0.00],[66.5,0.00],[67.5,0.20],[68.5,0.20],[69.5,0.20],[70.5,0.20],[71.5,0.40],[72.5,0.30],[73.5,0.30],[74.5,0.30],[75.5,0.10],[76.5,-0.10],[77.5,-0.20],[78.5,-0.40],[79.5,-0.40],[80.5,-0.20],[81.5,0.00],[82.5,0.00],[83.5,0.20],[84.5,0.20],[85.5,0.20],[86.5,-0.20],[87.5,-0.20],[88.5,-0.20],[89.5,0.00],[90.5,0.00],[91.5,0.00],[92.5,0.20],[93.5,0.20],[94.5,0.00],[95.5,0.00],[96.5,0.20],[97.5,-0.20],[98.5,-0.40],[99.5,-0.40],[100.5,-0.40],[101.5,-0.40],[102.5,-0.20],[103.5,0.00],[104.5,0.00],[105.5,0.00],[106.5,0.00],[107.5,-0.20],[108.5,-0.20],[109.5,-0.20],[110.5,-0.20],[111.5,-0.20],[112.5,-0.20],[113.5,-0.20],[114.5,-0.40],[115.5,-0.40],[116.5,-0.40],[117.5,-0.20],[118.5,0.00],[119.5,0.20],[120.5,0.40],[121.5,0.40],[122.5,0.60],[123.5,0.40],[124.5,0.40],[125.5,0.20],[126.5,0.20],[127.5,0.00],[128.5,0.00],[129.5,-0.20],[130.5,-0.20],[131.5,-0.20],[132.5,-0.20],[133.5,-0.20],[134.5,0.00],[135.5,0.20],[136.5,0.40],[137.5,0.40],[138.5,0.40],[139.5,0.40],[140.5,0.20],[141.5,-0.20],[142.5,-0.20],[143.5,-0.20],[144.5,-0.20],[145.5,-0.40],[146.5,0.00],[147.5,0.00],[148.5,0.00],[149.5,0.00],[150.5,0.00],[151.5,-0.40],[152.5,-0.40],[153.5,-0.40],[154.5,-0.40],[155.5,0.00],[156.5,0.20],[157.5,0.00],[158.5,0.00],[159.5,0.20],[160.5,0.00],[161.5,0.20],[162.5,0.40],[163.5,0.60],[164.5,0.40],[165.5,0.60],[166.5,0.20],[167.5,0.20],[168.5,0.00],[169.5,0.10],[170.5,-0.10],[171.5,-0.10],[172.5,-0.10],[173.5,-0.30],[174.5,-0.40],[175.5,-0.20],[176.5,0.00],[177.5,0.00],[178.5,0.20],[179.5,0.20],[180.5,0.00],[181.5,0.20],[182.5,0.40],[183.5,0.40],[184.5,0.40],[185.5,0.40],[186.5,0.20],[187.5,0.00],[188.5,0.00],[189.5,0.00],[190.5,0.00],[191.5,0.20],[192.5,0.20],[193.5,0.00],[194.5,0.00],[195.5,0.20],[196.5,0.00],[197.5,-0.20],[198.5,0.00],[199.5,0.00],[200.5,-0.20],[201.5,-0.10],[202.5,0.10],[203.5,-0.10],[204.5,-0.30],[205.5,-0.30],[206.5,-0.40],[207.5,-0.40],[208.5,-0.20],[209.5,-0.20],[210.5,-0.20],[211.5,-0.20],[212.5,-0.40],[213.5,-0.20],[214.5,0.00],[215.5,-0.20],[216.5,-0.20],[217.5,0.20],[218.5,0.10],[219.5,0.10],[220.5,0.30],[221.5,0.30],[222.5,0.10]];dis_datapoints = [];trans_datapoints = [];sec_helix_datapoints = [[8,10],[57,66],[79,85]];sec_strand_datapoints = [[13,22],[25,31],[41,49],[93,100],[104,113],[117,126],[148,152],[156,168],[172,183],[195,200],[201,203],[210,221]];acc_exposed_datapoints = [[1,6],[9,10],[13,13],[15,15],[19,19],[23,23],[26,26],[32,32],[34,35],[37,37],[39,39],[41,41],[50,51],[53,53],[70,70],[74,74],[77,78],[81,81],[84,84],[88,90],[92,92],[110,110],[114,117],[128,128],[131,132],[134,134],[137,140],[142,142],[144,145],[148,148],[154,155],[158,158],[168,169],[171,171],[178,178],[182,182],[184,186],[188,188],[191,191],[200,200],[203,203],[205,207],[209,209],[212,212],[215,216],[218,218],[223,225]];acc_buried_datapoints = [[7,8],[12,12],[14,14],[16,16],[18,18],[20,20],[22,22],[24,24],[27,27],[29,29],[31,31],[33,33],[40,40],[44,44],[46,46],[48,48],[54,58],[60,65],[67,68],[71,72],[79,79],[82,83],[87,87],[91,91],[93,93],[95,97],[99,99],[103,107],[111,111],[113,113],[118,118],[120,120],[122,122],[124,124],[126,126],[129,129],[133,133],[135,136],[146,146],[150,151],[156,157],[159,159],[161,161],[163,163],[165,165],[167,167],[173,173],[175,175],[177,177],[179,179],[183,183],[187,187],[189,190],[194,195],[197,197],[199,199],[211,211],[217,217],[219,219]];flot_plot_options = []; flot_plot_options[0] = {grid: {borderWidth: {top: 0,right: 0,bottom: 0,left: 0}},legend: {show: false},xaxes: [{show: true,min: 0,max: 200,ticks: [[0.5, '1'], [24.5, '25'], [49.5, '50'], [74.5, '75'], [99.5, '100'], [124.5, '125'], [149.5, '150'], [174.5, '175'], [199.5, '200']],tickLength: -5}],yaxes: [{show: true,ticks: [[0, '0'], [4.5,'hydro-<br>phobic '], [-4.5,'hydro-<br>philic ']],min: -4.5,max: +4.5,font: {size: 12,lineHeight: 14,style: 'italic',weight: 'bold',family: 'sans-serif',variant: 'small-caps',color: 'rgba(100,149,237,1)'}},{show: true,ticks: [[0, ''], [1,'positive<br> charge'], [-1,'negative<br> charge']],position: 'right',min: -1,max: 1,font: {size: 12,lineHeight: 14,style: 'italic',weight: 'bold',family: 'sans-serif',variant: 'small-caps',color: 'rgba(255,99,71,1)'}}]};number_of_plots = 2;for ( plot_num = 1 ; plot_num < number_of_plots ; plot_num ++){flot_plot_options[plot_num] = jqAutoAnnotator.extend(true, {} ,flot_plot_options[0]);flot_plot_options[plot_num].xaxes = [{min: plot_num*200,max: (plot_num + 1)*200,ticks: [ [plot_num*200 + 0.5, (plot_num*200 + 1).toString()], [plot_num*200 + 24.5, (plot_num*200 + 25).toString()], [plot_num*200 + 49.5, (plot_num*200 + 50).toString()], [plot_num*200 + 74.5, (plot_num*200 + 75).toString()], [plot_num*200 + 99.5, (plot_num*200 + 100).toString()], [plot_num*200 + 124.5, (plot_num*200 + 125).toString()], [plot_num*200 + 149.5, (plot_num*200 + 150).toString()], [plot_num*200 + 174.5, (plot_num*200 + 175).toString()], [plot_num*200 + 199.5, (plot_num*200 + 200).toString()] ],tickLength: -5}];};try {if( jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_button').val() =='Show' ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_container').css('display','block');jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_button').val('Hide');var description_html = '<div id=\'AutoAnnotator_plot_selectors\'>';description_html = description_html + '<br> <input type=\'checkbox\' id=\'hydrophobicity_checkbox\' checked=\'checked\'> Moving average over 5 amino acids for hydrophobicity (<img src=\'https://static.igem.org/mediawiki/2013/e/e9/TUM13_hydrophobicity_icon.png\' alt=\'blue graph\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'charge_checkbox\' checked=\'checked\'> Moving average over 5 amino acids for charge (<img src=\'https://static.igem.org/mediawiki/2013/3/3e/TUM13_charge_icon.png\' alt=\'red graph\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'dis_checkbox\' checked=\'checked\'> Predicted disulfid bridges (<img src=\'https://static.igem.org/mediawiki/2013/2/28/TUM13_dis_icon.png\' alt=\'yellow circle\' height=\'10\'></img>) with the number of the bridge in the center';description_html = description_html + '<br> <input type=\'checkbox\' id=\'trans_checkbox\' checked=\'checked\'> Predicted transmembrane helices (<img src=\'https://static.igem.org/mediawiki/2013/7/78/TUM13_trans_icon.png\' alt=\'turquois bars\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'sec_checkbox\' checked=\'checked\'> Predicted secondary structure: Helices (<img src=\'https://static.igem.org/mediawiki/2013/b/bf/TUM13_helix_icon.png\' alt=\'violet bars\' height=\'10\'></img>) and beta-strands (<img src=\'https://static.igem.org/mediawiki/2013/b/bf/TUM13_strand_icon.png\' alt=\'yellow bars\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'acc_checkbox\' checked=\'checked\'> Predicted solvent accessability: Exposed (<img src=\'https://static.igem.org/mediawiki/2013/1/16/TUM13_exposed_icon.png\' alt=\'blue bars\' height=\'10\'></img>) and buried (<img src=\'https://static.igem.org/mediawiki/2013/0/0b/TUM13_buried_icon.png\' alt=\'green bars\' height=\'10\'></img>) residues';description_html = description_html + '<br></div>';jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_explanation').html(description_html);plot_according_to_selectors_1397080705152();jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #AutoAnnotator_plot_selectors').find('input').click(plot_according_to_selectors_1397080705152);}else{jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_container').css('display','none');jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_button').val('Show');jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_explanation').html('');}}catch(err){txt='There was an error with the button controlling the visibility of the plot.\n';txt=txt+'The originating error is:\n' + err + '\n\n';alert(txt);}};function plot_according_to_selectors_1397080705152(){try{var plot_datasets = [[],[]];if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_checkbox').prop('checked') == true){plot_datasets[0] = { color: 'rgba(100,149,237,1)',data: hydrophobicity_datapoints,label: 'Hydrophobicity',lines: { show: true, fill: true, fillColor: 'rgba(100,149,237,0.1)' },yaxis: 1};}if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #charge_checkbox').prop('checked') == true){plot_datasets[1] = {color: 'rgba(255,99,71,1)',data: charge_datapoints,label: 'Charge',lines: { show: true, fill: true, fillColor: 'rgba(255,99,71,0.1)' },yaxis: 2};}for (plot_num = 0 ; plot_num < number_of_plots ; plot_num ++){jqAutoAnnotator.plot('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder'+ plot_num.toString(), plot_datasets, flot_plot_options[plot_num] );}var screen_width = jqAutoAnnotator('canvas.flot-base').width(); var pos_of_first_tick = 46;var pos_of_last_tick = screen_width - 51;var tick_diff = (screen_width - 97)/199;if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #dis_checkbox').prop('checked') == true){for ( j = 0 ; j < dis_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((dis_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_dis\' style=\'left:' + ((pos_of_first_tick - 8 + (dis_datapoints[j][0] - 1)*tick_diff - Math.floor((dis_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px;\'><b>' + (j+1) + '</b></div>');jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((dis_datapoints[j][1] - 1)/200) ).append('<div class=\'AutoAnnotator_dis\' style=\'left:' + ((pos_of_first_tick - 8 + (dis_datapoints[j][1] - 1)*tick_diff - Math.floor((dis_datapoints[j][1] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px;\'><b>' + (j+1) + '</b></div>');}}if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #trans_checkbox').prop('checked') == true){for ( j = 0 ; j < trans_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((trans_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_trans\' style=\'width:' + (((trans_datapoints[j][1] - trans_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px ;left:' + ((pos_of_first_tick + (trans_datapoints[j][0] - 1.5)*tick_diff - Math.floor((trans_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #sec_checkbox').prop('checked') == true){for ( j = 0 ; j < sec_helix_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((sec_helix_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_sec_helix\' style=\'width:' + (((sec_helix_datapoints[j][1] - sec_helix_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (sec_helix_datapoints[j][0] - 1.5)*tick_diff - Math.floor((sec_helix_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}for ( j = 0 ; j < sec_strand_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((sec_strand_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_sec_strand\' style=\'width:' + (((sec_strand_datapoints[j][1] - sec_strand_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (sec_strand_datapoints[j][0] - 1.5)*tick_diff - Math.floor((sec_strand_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}if(jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #acc_checkbox').prop('checked') == true){for ( j = 0 ; j < acc_buried_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((acc_buried_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_acc_buried\' style=\'width:' + (((acc_buried_datapoints[j][1] - acc_buried_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (acc_buried_datapoints[j][0] - 1.5)*tick_diff - Math.floor((acc_buried_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}for ( j = 0 ; j < acc_exposed_datapoints.length ; j++ ){jqAutoAnnotator('#AutoAnnotator_container_1397080705152 #hydrophobicity_charge_placeholder' + Math.floor((acc_exposed_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_acc_exposed\' style=\'width:' + (((acc_exposed_datapoints[j][1] - acc_exposed_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (acc_exposed_datapoints[j][0] - 1.5)*tick_diff - Math.floor((acc_exposed_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}}catch(err){txt='There was an error while drawing the selected elements for the plot.\n';txt=txt+'The originating error is:\n' + err + '\n\n';throw(txt);}}</script></html> | ||
Revision as of 22:07, 25 August 2020
**highly** engineered mutant of red fluorescent protein from Discosoma striata (coral)
monomeric RFP: Red Fluorescent Protein. Excitation peak: 584 nm Emission peak: 607 nm
Usage and Biology
Robert E. Campbell started with Discosoma RFP (DsRed) and evolved a faster folding, monomeric variant. See paper listed in source. Codon optimized for expression in bacteria (?? DE)
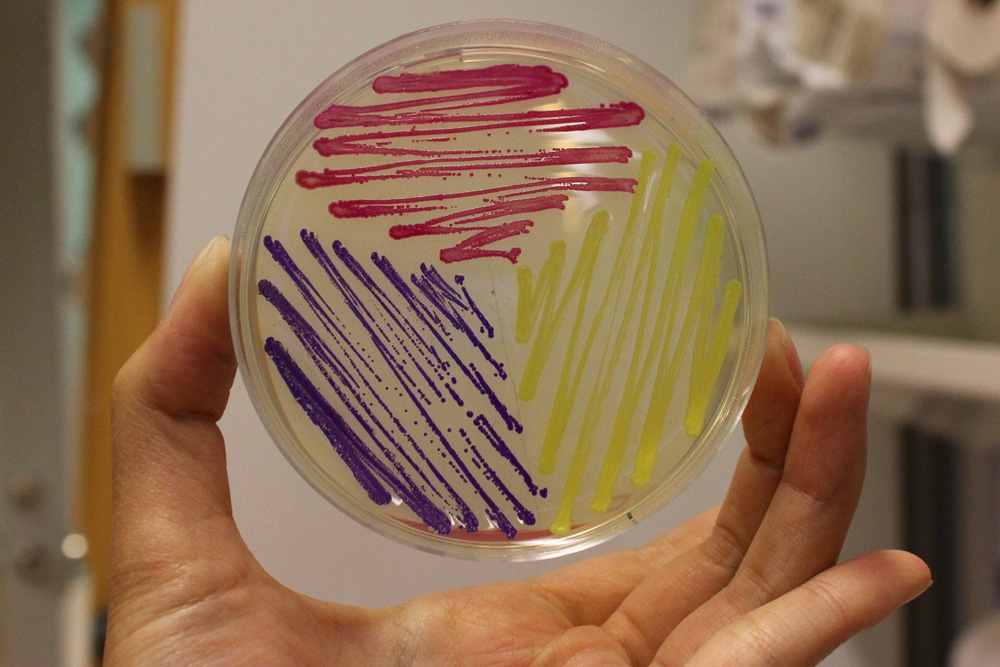
iGEM11_Uppsala-Sweden: Expression of chromoproteins. The images above show E coli No part name specified with partinfo tag. expressing amilCP BBa_K592009 (blue), amilGFP BBa_K592010 (yellow) and RFP BBa_E1010 (red).
Peking iGEM 2016 has fused this part with triple spytag. The fused protein is participate in Peking’s polymer network. By adding this protein, the whole polymer network become visible in most conditions. If you want to learn more about Peking’s polymer network and the role of mRFP in this network, please click here https://parts.igem.org/Part:BBa_K1989004".
Allergen characterization of BBa_E1010: NOT a potential allergen
The Baltimore Biocrew 2017 team discovered that proteins generated through biobrick parts can be evaluated for allergenicity. This information is important to the people using these parts in the lab, as well as when considering using the protein for mass production, or using in the environment. The allergenicity test permits a comparison between the sequences of the biobrick parts and the identified allergen proteins enlisted in a data base.The higher the similarity between the biobricks and the proteins, the more likely the biobrick is allergenic cross-reactive. In the full-length alignments by FASTA, 30% or more amount of similarity signifies that the biobrick has a Precaution Status meaning there is a potential risk with using the part. A 50% or more amount of identity signifies that the biobrick has a Possible Allergen Status. In the sliding window of 80 amino acid segments, greater than 35% signifies similarity to allergens. The percentage of similarity implies the potential of harm biobricks’ potential negative impact to exposed populations. For more information on how to assess your own biobrick part please see the “Allergenicity Testing Protocol” in the following page http://2017.igem.org/Team:Baltimore_Bio-Crew/Experiments
For the biobrick part, BBa_E1010, there was a 27.6% of identity match and 56.9% of similarity match compared to the allergen database. This means that the biobrick part is not of potential allergen status. In the 80 amino acid alignments by FASTA, no matches found that are greater than 35% for this biobrick.
>Internal Priming Screening Characterization of BBa_E1010: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
Contribution
Group: USAFA iGEM 2019
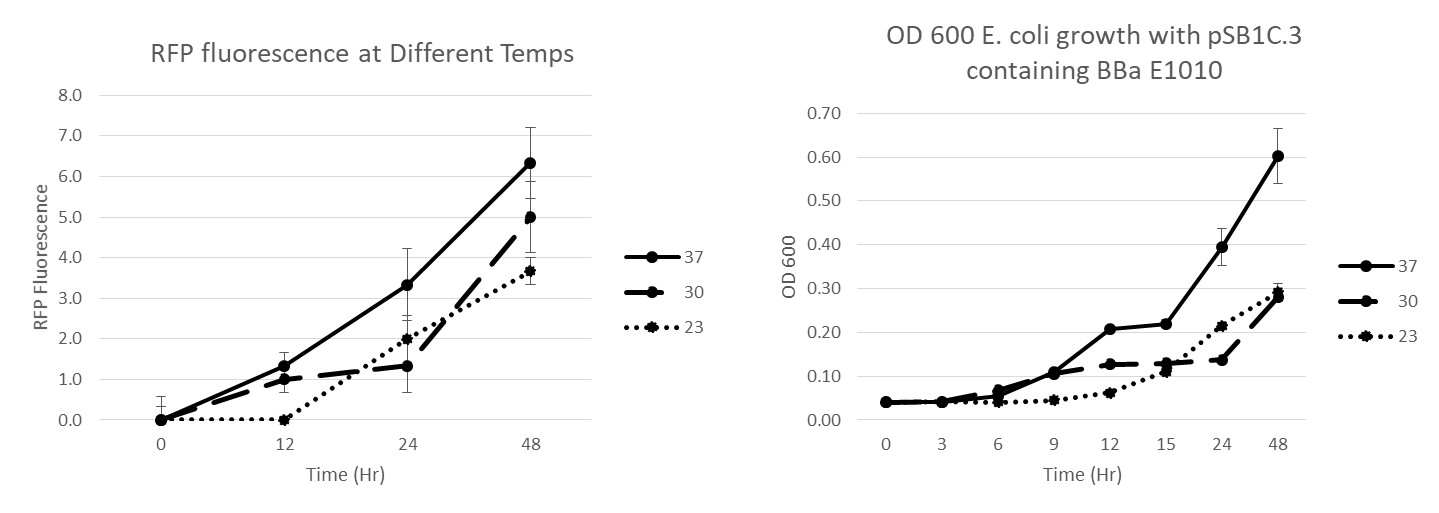
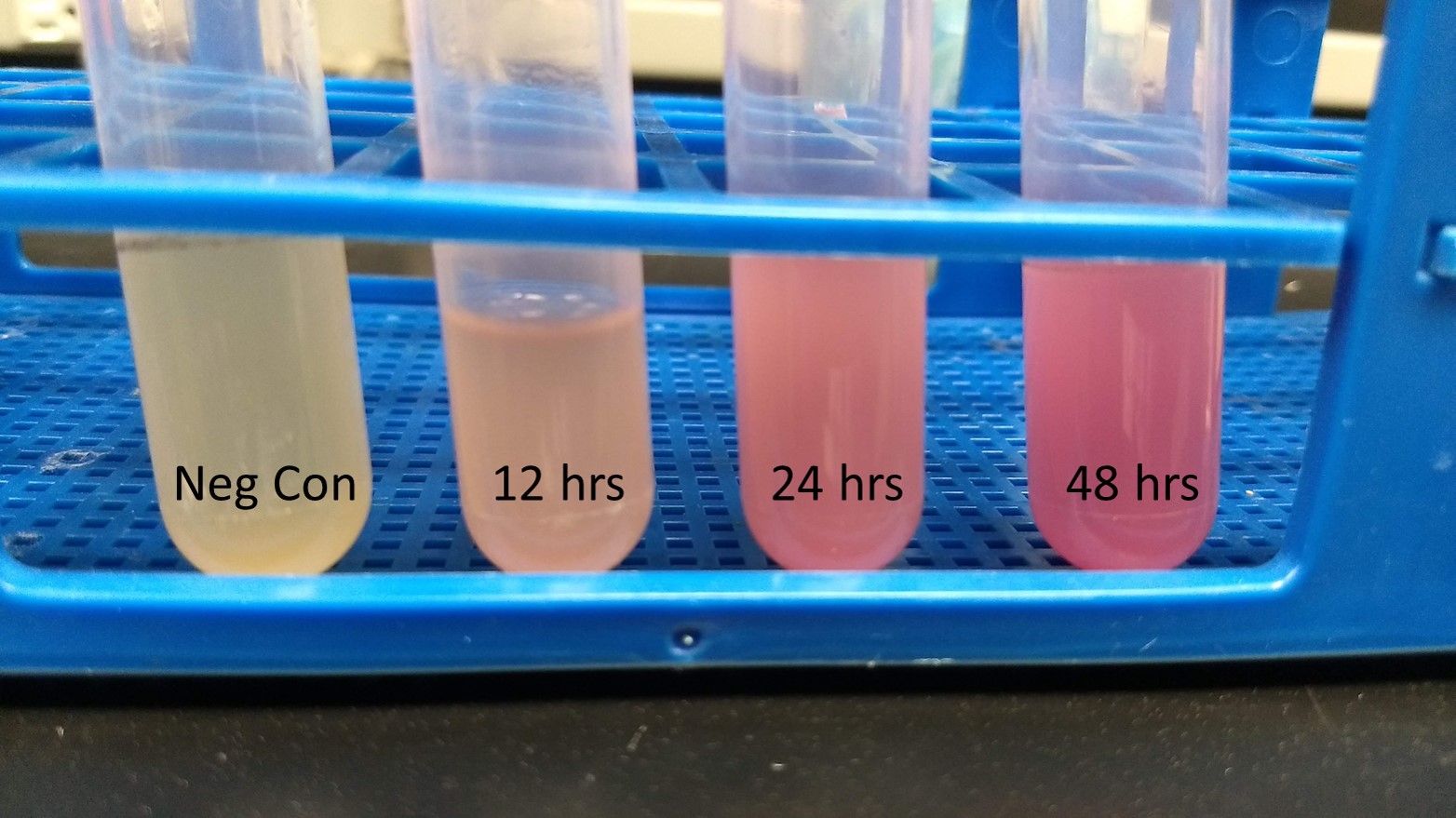
Summary: We have characterized the fluoresence of this part over time and at different temperatures in order to determine optimal incubation conditions and times for best mRFP expression. Our results indicate that 37 degrees C is optimal for both growth and mRFP expression. At 12 hours of incubation at 37C, the RFP was detectable, but after 24 and 48 hours, the expression was much more robust.
Conclusion: BBaE1010 is a strong reporter that has RFP expression that can be detected with a fluorescent plate reader or visualized by the unaided eye. 37C was the optimal growth temperature for E. coli expressing the BBaE1010 mRFP and the expression/detection of the mRFP increased over time with 48 hours showing the most robust red color.
Pre-experiment
E. coli Nissle 1917 strains were grown overnight in Lysogeny Broth (LB) containing ampicilin (100 µg/mL) at 37°C and 200 rpm. Cultures were diluted in fresh LB until achieve 0,1 OD with the corresponding antibiotic and transferred to a 96-well plate (50 µL/well). Samples were always made in triplicates and a blank of LB. During 8h the absorbance at OD600 and fluorescence (excitation 488 nm and emission 530 nm) were measured with intervals of 1 hour.
Contribution
Group: USAFA iGEM 2019
Summary: We have characterized the fluoresence of this part over time and at different temperatures in order to determine optimal incubation conditions and times for best mRFP expression. Our results indicate that 37 degrees C is optimal for both growth and mRFP expression. At 12 hours of incubation at 37C, the RFP was detectable, but after 24 and 48 hours, the expression was much more robust.
Conclusion: BBaE1010 is a strong reporter that has RFP expression that can be detected with a fluorescent plate reader or visualized by the unaided eye. 37C was the optimal growth temperature for E. coli expressing the BBaE1010 mRFP and the expression/detection of the mRFP increased over time with 48 hours showing the most robust red color.
Contribution
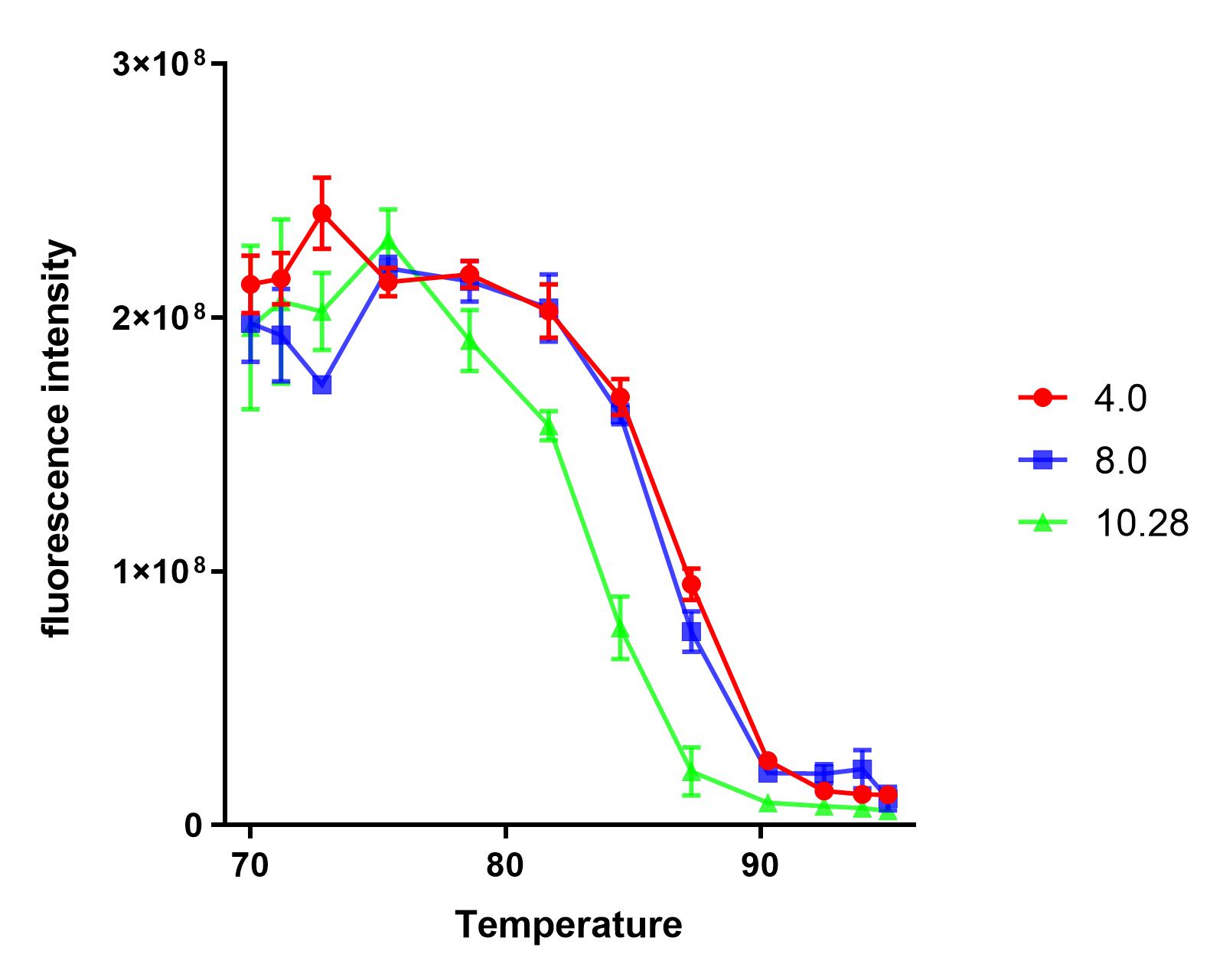
Group:NAU_CHINA 2019
Summary: We measured the fluorescence signal of mRFP in buffer under different pH values and temperatures.
Report genes are essential for every team to construct genetic pathways. We believe that every team has a J04450 complex. After all, it is our favorite report gene!
This year, the NAU iGEM characterized E1010. Based on the work of the previous team work, we explored the tolerance of E1010 expression products at different pH values and temperatures.
Pre-experiment
We dispensed a small amount of the sample into 12 PCR tubes, and a gradient test at 65-95℃ was conducted in a PCR machine for 10 minutes.
We found that the protein samples showed significant denaturation when the temperature was higher than 75℃. So, we set the experimental temperature later at and above 75℃.
The purified sample was dispensed into four PCR tubes and the pH was) adjusted to 4.0, 8.0, 10.28, respectively.
Under each pH value, we dispensed each sample into 12 PCR tubes(50uL sample + 50uL buffer), and a gradient test at 75-95℃ was conducted in a PCR machine for 10 minutes.
We measured the fluorescence intensity of the product under the microplate reader (excitation wavelength was 584 nm, and absorption wavelength was 611 nm).

Analysis
The results show that the product of J04450 begins to show significant denaturation at 84.4℃, and the fluorescence value of the protein no longer changes significantly at the temperature of around 92.5℃, which means E1010 product is completely denatured (shown in our characterization map with a small amount of protein precipitation and the color is pale yellow).
The temperature (from 75.4℃ to 90.3℃) and fluorescence values show a significant linear relationship .
At the same temperature, the pH=4.0 and pH=8.0 curves were basically consistent. The stability of the E1010 expression products at these two pH values was significantly better than that under the condition of pH=10.28.
Discussion
Report genes are required for the construction of most gene pathways. In the future iGEM journey, other teams and we may use some bacteria, like thermophiles, that grow in extreme conditions. We hope we can still use our familiar report gene in these bacteria to detect protein secretion and other related circumstances, so we conducted these tests.
We simulated the extreme environments in which we released the expression products to the extracellular—acid, weak acid, alkaline, and high temperatures—by adjusting the temperature gradients and pH values.
We hope that our experimental results can provide other teams with a reference for the stability of report gene expression products in extreme environments.
Reference
[1] Sudhagar S A , Prasad K P , Makesh M , et al. Characterization and production of polyclonal antisera against pangasius (Pangasianodon hypophthalmus) serum immunoglobulin IgM derived from DEAE cellulose based ion exchange chromatography[J]. Aquaculture Research, 2015, 46(6):1417-1425.
Contribution
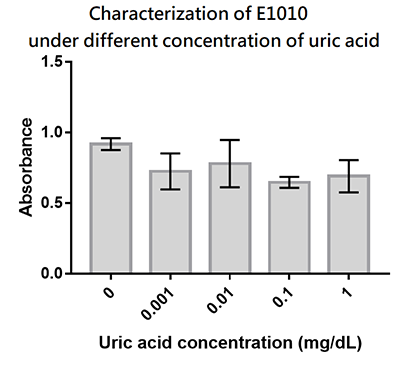
Group: Hong_Kong_LFC_PC 2019
Summary: We designed a uric-acid-sensitive uricase generator for treating the gout disease. Since we will use the fluorescence protein to track the gene expression of the device, we would like to check the effect of uric acid on the signal of this E1010 RFP. Since uric acid can alter the pH value, it may interfere the folding of RFP and lower its fluorescence signal. We treat the extracted RFP with different concentration of uric acid, and found that uric acid has a very little effect on the fluorescence signal. This indicate that our system is possible to track the gene expression.
Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We have adapted the part to be able to assemble transcriptional units with the Golden Gate method and we have done the characterization of this protein.
Documentation:
Part BBa_K2656014 is the monomeric Red Fluorescent Protein 1 coding sequence BBa_E1010 adapted into the [http://2018.igem.org/Team:Valencia_UPV/Design Golden Braid assembly method]. Thus, this sequence is both compatible with the BioBrick and GoldenBraid 3.0. grammar.
This coding sequence can be combined with other Golden Braid compatible parts from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Valencia UPV IGEM 2018 Printeria Collection] to assemble transcriptional units in a one-step BsaI reaction with the [http://2018.igem.org/Team:Valencia_UPV/Protocols Golden Gate assembly protocol].
The characterization of this protein (and by extension of all the other part that codify for the mRFP1) was performed with our transcriptional unit BBa_K2656109. This transcriptional unit was assembled in a GoldenBraid alpha1 plasmid including the following parts:
- BBa_K2656004: the J23106 promoter in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- BBa_K2656009: the B0030 ribosome biding site in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- BBa_K2656014: coding sequence
- BBa_K2656026: the B0015 transcriptional terminator in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
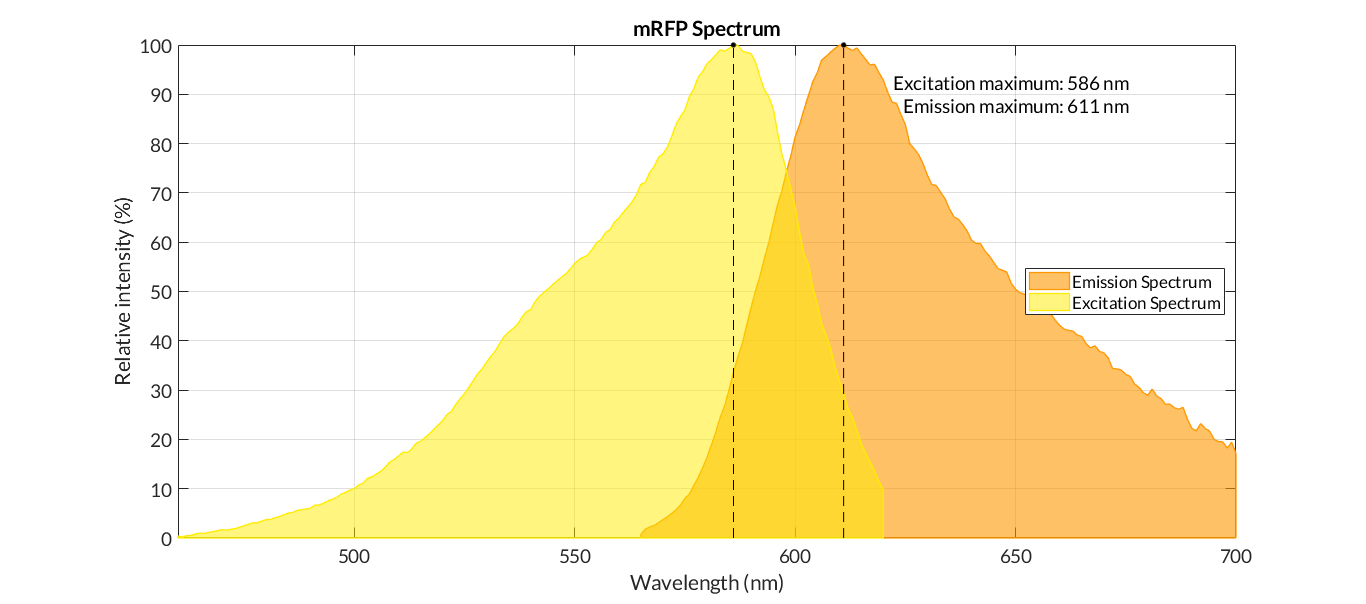
In order to carry out a correct characterization of the protein and to be able to use it to make measurements of the different transcriptional units that we assembled with it, we obtained the emission and excitation spectra in the conditions of our equipment. By using this protocol [http://2018.igem.org/Team:Valencia_UPV/Experiments#spectra] with the parameters of Table 1, Figure 1 was obtained.
| Parameter | Value | ||
| Number of samples | 3 | ||
| Excitation Wavelength measurement range 1 (nm) | [450-620] | ||
| Excitation Wavelength measurement range 2 (nm) | [620-700] | ||
| Emission wavelenght 1 (nm) | 650 | ||
| Emission wavelenght 2 (nm) | 590 | ||
| Emission Wavelength measurement range (nm) | [565-700] | ||
| Excitation wavelenght (nm) | 540 | ||
| Gain (G) | 70 | ||
| Table 1. Parameters used to obtain the spectra | |||
Contribution
Group: Hong Kong-CUHK iGEM 2017
Author: Yuet Ching Lin
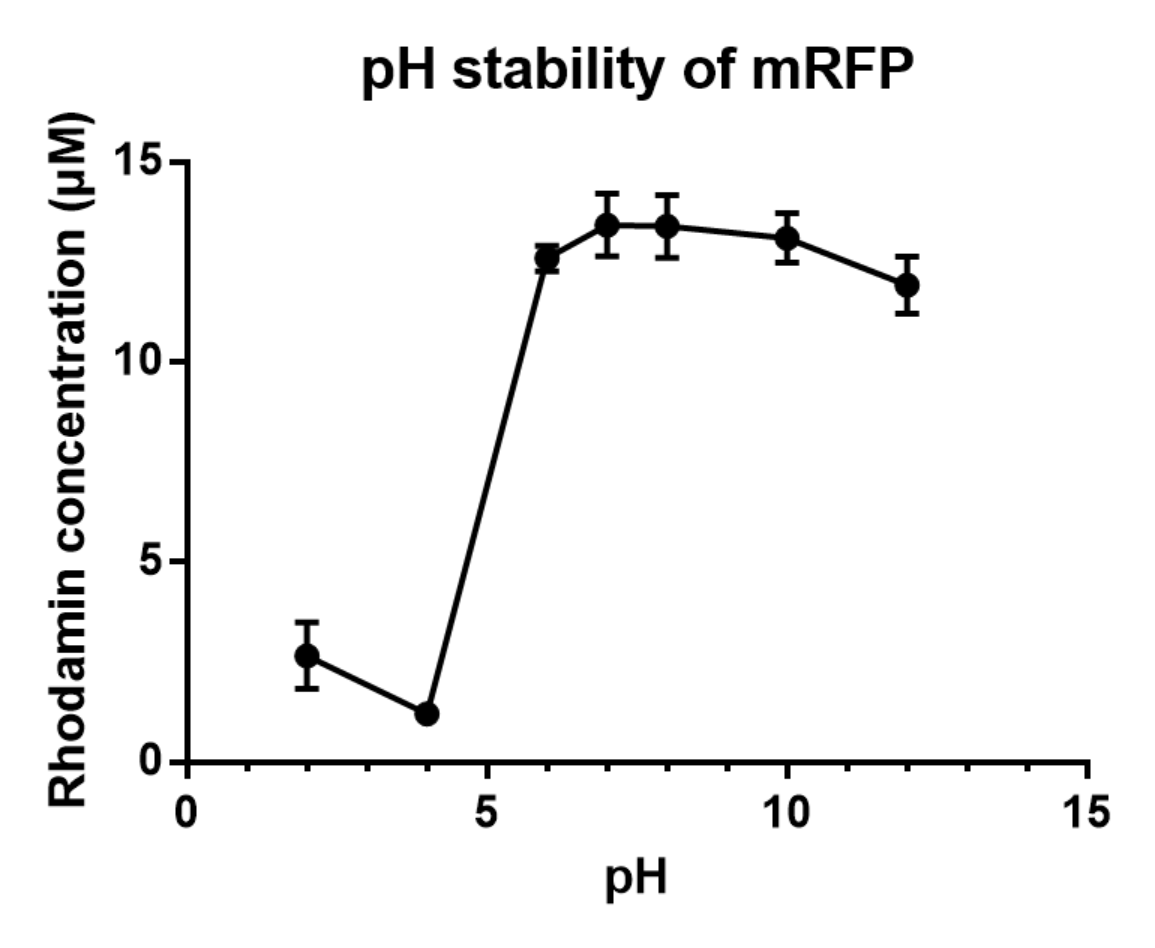
Summary: We measured the fluorescent signal of mRFP in buffers with different pH.
Documentation:
Charaterization of mRFP pH stabillity:
We grew C41 bacteria with parts BBa_J61002 in 2XYT for 24 hours. After purifying the mRFP by Ion Exchange Chromatography and Hydrophobic Interaction Chromatography, we measured the fluoresece (ex ,em ) of purified mRFP, which is diluted to 10µg/100µl (total 200µl) in triplicates, into different buffers (ranges from pH2 to pH12; Volume of mRFP:buffer = 1:1.8). To facilitate reproducibility of the experiment, we correlated the relative fluorescent intensity to an absolute fluorophore concentration by referring it to a standard curve of the fluorophores(Rhodamine) using the interlab study protocol. The result shows that the stability drops dramatically in pH condition below 6 and relatively stable in pH 6-10.
| Measurement Type | Fluorescence |
| Microplate name | COSTAR 96 |
| Scan mode | orbital averaging |
| Scan diameter [nm] | 3 |
| Excitation | 550-20 |
| Emission | 605-40 |
| Dichronic filter | auto 572.5 |
| Gain | 500 |
| Focal height [nm] | 9 |
Contribution2
Part name:BBa_K2382013
Group: iGEM17_CSMU_NCHU_Taiwan 2017
Author: SHAO-CHI LO
Summary: We add a His Tag at the end. Therefore, a fusion protein with this part may have red color and the ability to be purified easily.
Documentation:
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 555
Illegal AgeI site found at 667 - 1000COMPATIBLE WITH RFC[1000]
Parts table
| Protein data table for BioBrick BBa_E1010 automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 10: (underlined part encodes the protein) ATGGCTTCC ... ACCGGTGCTTAATAACGCTGATAGTGCTAGTGTAGATCGC ORF from nucleotide position 1 to 675 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
Alignments (obtained from PredictProtein.org)
| ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
Subcellular Localization (reliability in brackets)
| Gene Ontology (reliability in brackets)
| |||||||||||||||||||||||||||||||||||||||||||||
Predicted features:
| ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.