Part:BBa_B0030
RBS.1 (strong) -- modified from R. Weiss
Strong RBS based on Ron Weiss thesis. Strength is considered relative to BBa_B0031, BBa_B0032, BBa_B0033 and BBa_B0034.
IIT Madras 2016's Characterization
Modelling
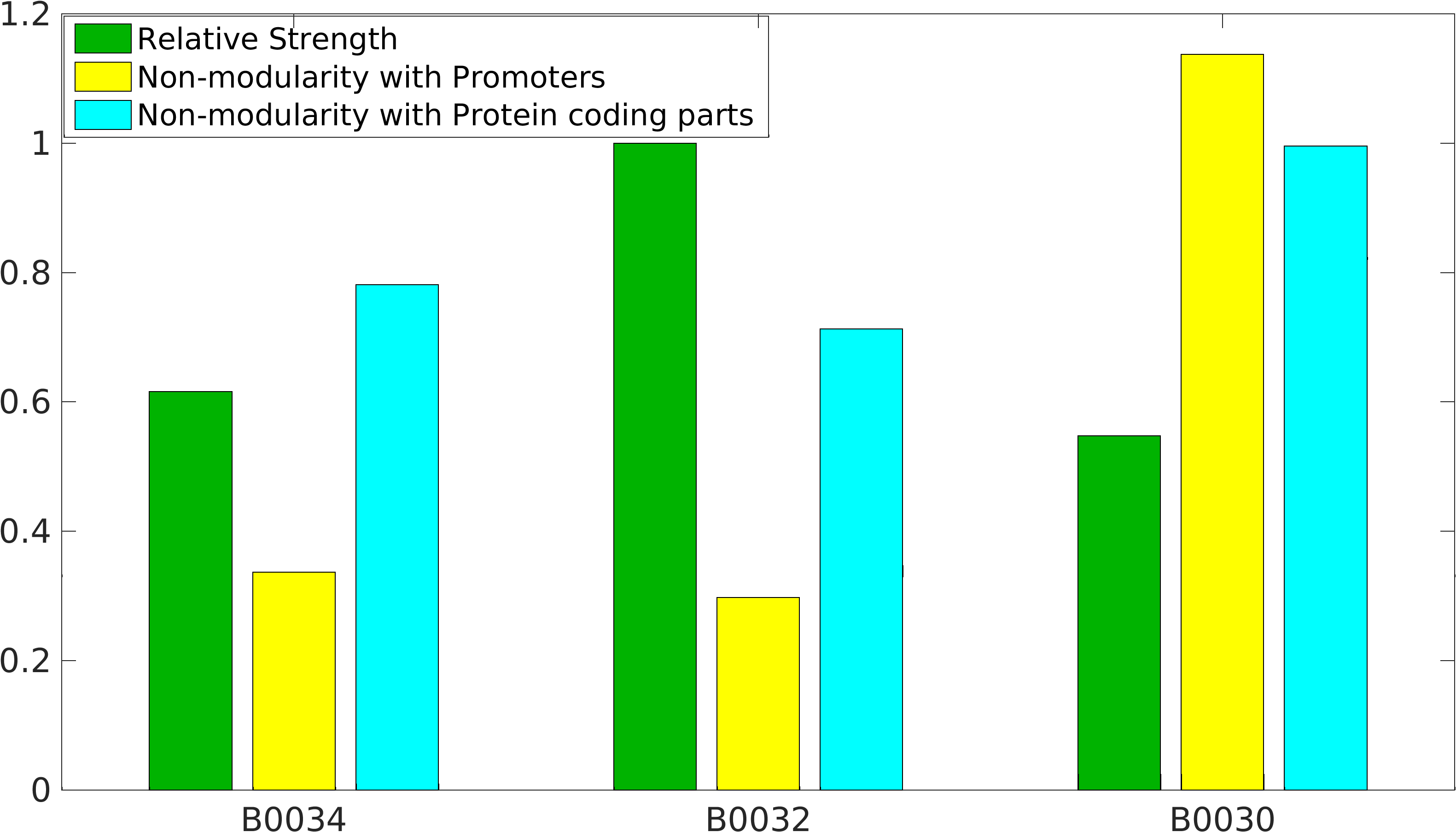
Global non-modularity towards promoters & protein coding parts and relative strength was estimated for RBSs B0030, B0032, B0034 in our [http://2016.igem.org/Team:IIT-Madras/Model#Modularity_of_RBS_parts modelling]
Experimentation
Biobrick RBSs B0030, B0031, B0032, B0034 were used in our 'Noise in Device' experiment to understand the role of RBS parts in giving rise to noise.
Note: not compatible with R0053 promoter due to likely transcript secondary structure This combination yielded very low gfp expression (see BBa_I7108). [jcb 8/3/05].
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Team Warsaw 2010's measurement
RBS strength (relative to B0034): 91,84%Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We have adapted the part to be able to assemble transcriptional units with the Golden Gate method and we have done the characterization of this RBS.
Documentation:
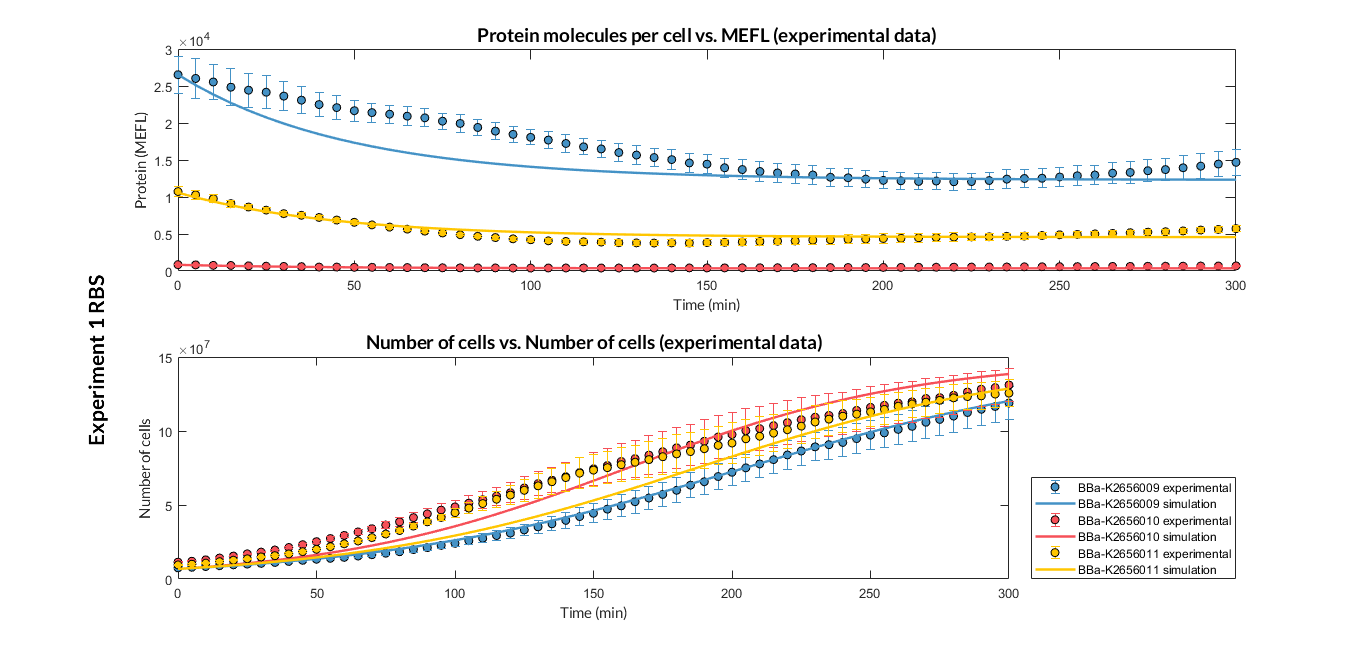
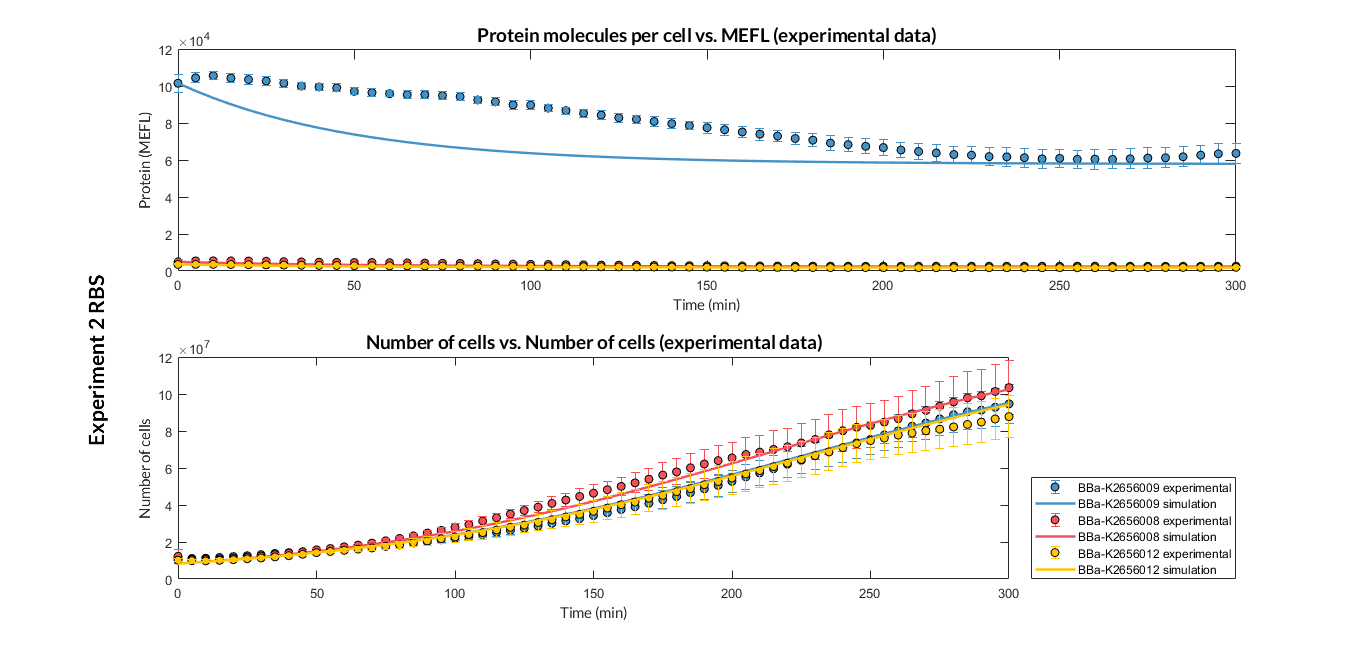
BBa_K2656009 is the BBa_B0030 RBS standardized into the Golden Braid assembly method. Thus, it is both compatibe with the BioBrick and Golden Gate grammar. It also includes the BioBrick equivalent scar in the 3' extreme, so the insertion of this supplementary bases ensure correct spacing for the CDS expression when assembled into a TU.
Characterization of the this part was performed with the transcriptional unit BBa_K2656101, which was used in two comparative RBS expression experiments with composite parts BBa_K2656102, BBa_K2656103 and BBa_K2656100, BBa_K2656104. They all were assembled in a Golden Braid alpha1 plasmid using the same promoter, CDS and terminator.
By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model]and rationale choose its optimized values based for each RBS tested.
We have calculated the relative strenght between the different RBS, taking BBa_K2656009 strong RBS as a reference. It has been defined as the quotient between the values of the protein in equilibrium of the results of the simulation of one RBS and the reference RBS. Likewise, a ratio between p parameters of the different RBS parts and p parameter of the reference RBS has been calculated.
| Table 2. BBa_K2656008 (GB BBa_B0032 RBS) relative strength and p ratio. | |||
| Parameter | Value | ||
| Relative strength | 1 | ||
| p parameter ratio (pRBS/pref) | 1 | ||
>Internal Priming Screening Characterization of BBa_B0030: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
Contribution: QHFZ-China 2019
Group: QHFZ-China iGEM 2019
Author: Cheng Li
Introduction & Design:
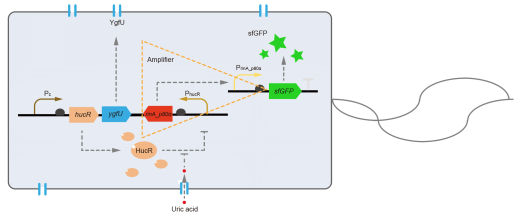
This year we use BBa_B0030 to construct BBa_K3007010 (https://parts.igem.org/Part:BBa_K3007010). We designed a gene circiut that can expressed sfGFP or dsRed under the induction of uric acid. RBS0030 (BBa_B0030) is a strong RBS and we used it to powerfully promote the expression of fluorescent proteins.
Documentation:
We charactered RBS0030 part in the part BBa_K3007010 (https://parts.igem.org/Part:BBa_K3007010). Fig.4 and Fig. 5 showed the result. Prin_p80α and RBS0030 controled the expression of sfGFP. Fig. 1-3 showed the process that we constructed the part.
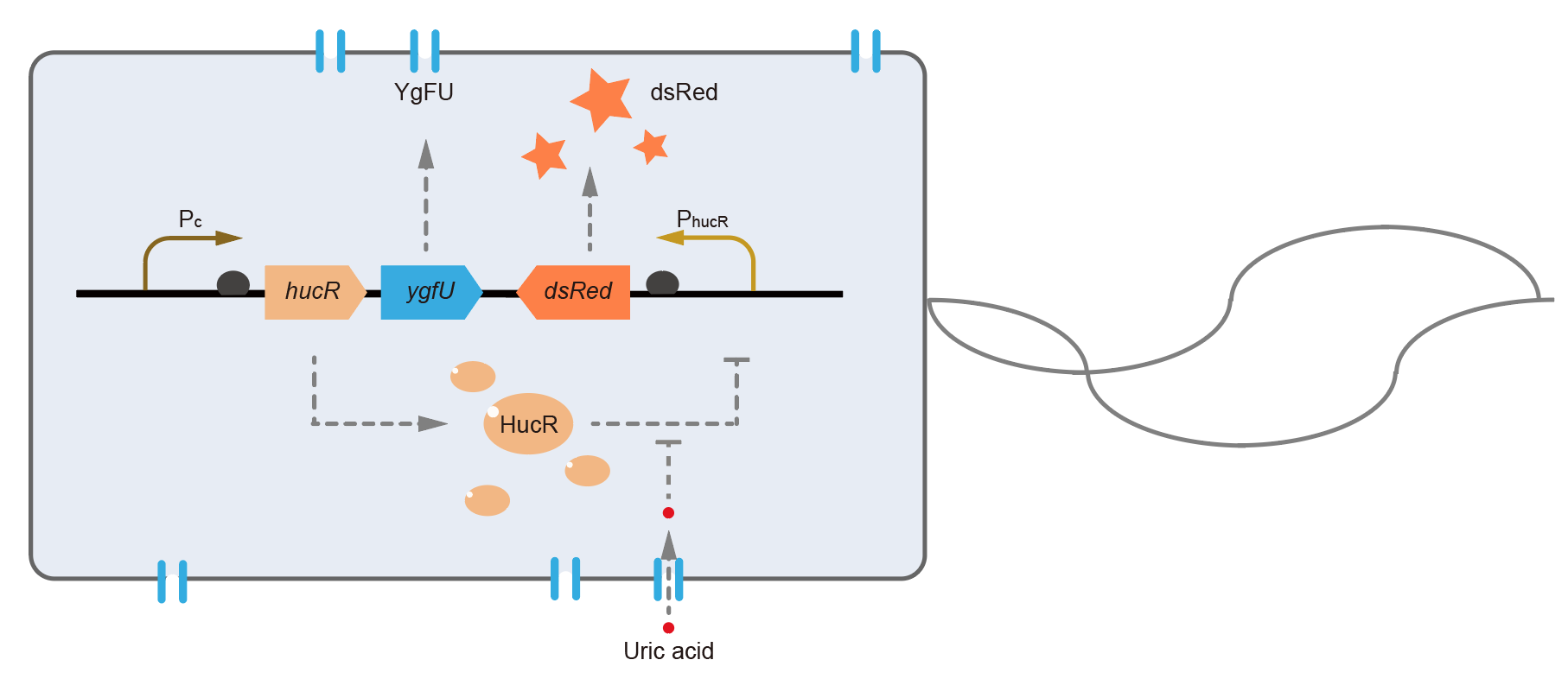
This year, QHFZ-China designed a UA monitor system in Escherichia coli (E. coli). The original version is shown in Fig. 1. Pc is a constitutive promoter, Pcp6 promoter, and it promotes the expression of HucR and YgfU. When uric acid is absent, HucR can bind to PhucR, which suppresses dsRed expression. If uric acid presents in high concentration, HucR will release from PhucR and the expression of dsRed will recover from the inhibition [2].
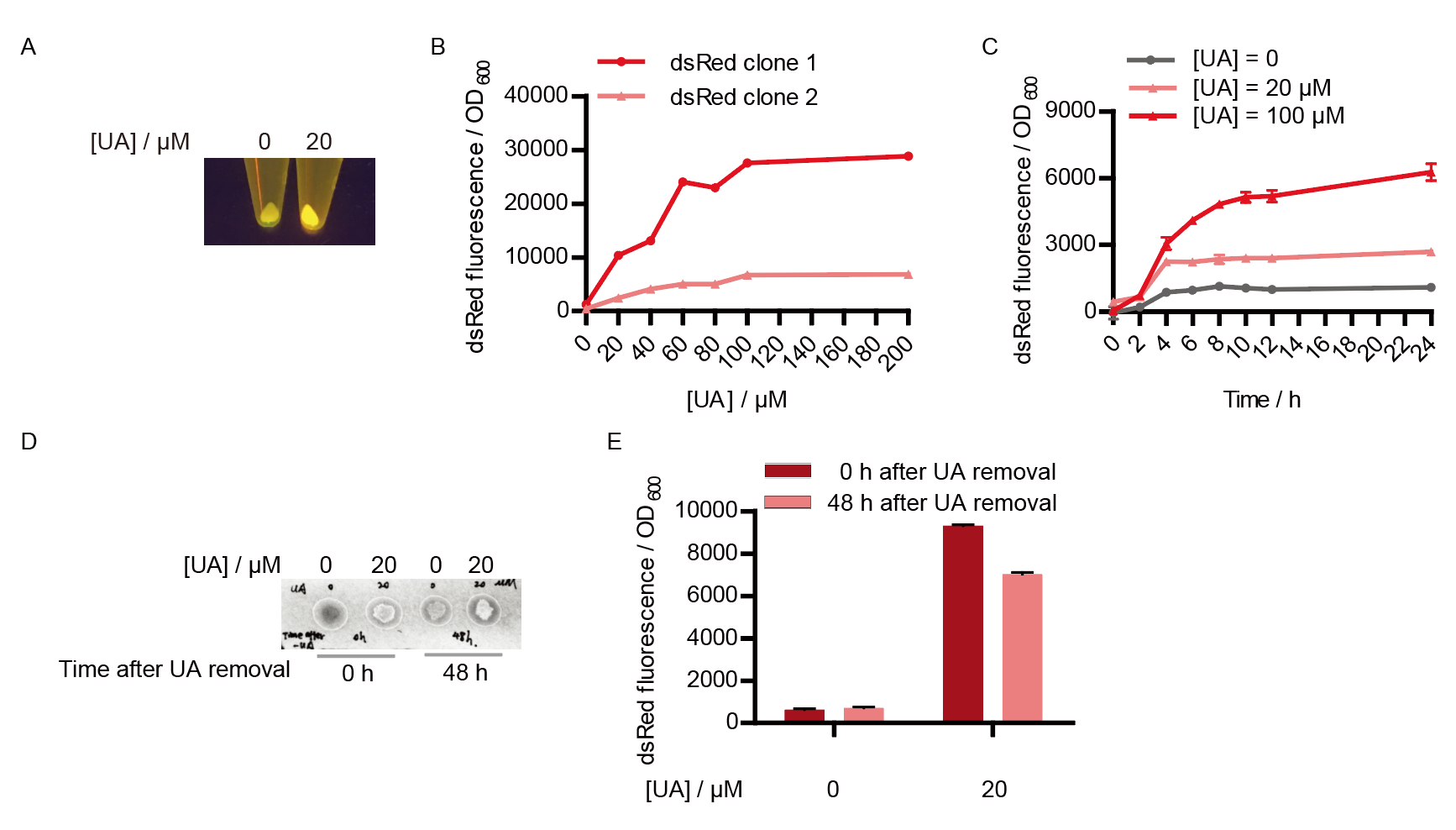
Two clones with the UA detection system were tested. The original gene circuit was able to response to UA in a range of 0 to 200 μM (Fig. 2A). The clone 1 showed much better dynamics than the other (Fig. 2B). Time course experiments showed that the fluorescence intensity became quite strong at 4 to 6 hours after UA induction, and became stable at 10 to 12 hours (Fig. 2C). Even if we removed UA by replacing fresh LB medium, after 48 hours shaking, the fluorescence would be still notable (Fig. 2D) and there was not significant difference of dsRed fluorescence / OD600 between before and after UA removing (Fig. 2E). All the data meant our design could detect high UA concentration quickly and stably.
However, through our human practices, we found the sensitivity and responding time of the original design are not good enough. In the next generation design, we introduced RinA_p80α - PrinA_p80a system to enhance the sensitivity. Meanwhile, we changed dsRed to sfGFP, whose maturation time is much shorter, to shorten the waiting time. The new version of the uric acid detector was shown in Fig. 3. If UA presented, RinA_p80α would express and active transcription of sfGFP which was under control of PrinA_p80α. We called this as Version 2.
We tested the sfGFP production of Version 2 under different concentration of extracellular UA. The curve in Fig. 4A showed the fluorescence was saturated under only 15 μM UA induction, while the old version needed about 100 μM UA to get saturated (Fig. 2B). To test if sfGFP could shorten the reaction time, we used the same construct only except reporter genes, called PRinA_p80α – sfGFP and PRinA_p80α – dsRed, respectively. After adding 20 μM UA into the reaction system, the curve of PRinA_p80α – sfGFP climbed much faster than PRinA_p80α – dsRed, which suggested our new design had a great induction performance, and fitted our predictions very well (Fig. 4B).

At last, we took a photo to show the green fluorescence released by E. coli expressing sfGFP.
References:
[1] Wan, X., Volpetti, F., Petrova, E., French, C., Maerkl, S. J., & Wang, B. (2019). Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nature chemical biology, 15(5), 540.
[2] Liang C., Xiong D., Zhang Y., Mu S. and Tang S. (2015). Development of a novel uric-acid-responsive regulatory system in Escherichia coli. Appl. Microbiol. Biotechnol. 99, 2267–2275.
Contribution: WHU-China 2019
pFadD_Lac is a fatty acid-sensitive promoter, one of the regulators in the enzymes of fatty acid biosynthesis in E. coli. It is composed of two fadR recognition sites. In the absence of fatty acids, an intrinsic transcription factor FadR acting as the repressor in the fatty acid metabolism system in E. coli. will attach to the promoter, and block the downstream gene expression. This has been proved to be effective by a previous iGEM team(NTHU_Taiwan). [1][2] 2019 NTHU_Taiwan further modified pFadD promoter by replacing its CRP binding site with a LacI repressor binding site, which reduced expression leakage. We combine the RBS(B0030) with the promoter pFadD_Lac which is sensitive to fatty-acids and use GFP to test the performance of the promotor by measuring the fluorescence intensity.

Figure 2. The change in sequence.
After the successful transformation of the plasmid, we used different concentrations of fatty acids for induction for different times. The expression level of eGFP was determined by fluorescence detection with a microplate reader. The results are as follows:

Figure 3. BBa_K3763002(pFadD_Lac-RBS BBa_B0030-GFP-rrnB T1 and T2 terminator, pDSW208-99-2) contains RBS without modification.

Figure 4. BBa_K3763003(pFadD_Lac-RBS BBa_K3763000-GFP-rrnB T1 and T2 terminator, pDSW208-99-2) Contains RBS with modification
Reference [1] https://2019.igem.org/Team:NTHU_Taiwan/Results [2] http://2012.igem.org/Team:NTU-Taida#
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
B0030 vs B0034 in E.coli BL21 (DE3) - Thessaly 2024
Group: Thessaly 2024
Purpose
BBa_B0030 and BBa_B0034 were the RBSes that we used in our promoter testing experiments. We wanted to check which RBS would be the strongest and which would be the ideal candidate for our design.
Methods
Measurements for OD 600nm and fluorescence (488nm,515nm) were taken over the course of 16 hours, in a 96-well microplate. Clonings were done according to the Golden Braid method, leaving us with level α constructs in the pDGB3a1 backbone ( BBa_K4213058. )
Constructs and pDGB3a1 (empty vector) transformed into E.coli BL21 (DE3) chassis, incubated at 37oC, 180rpm for 16 hours.
Here you can find more information about the constructs tested containing BBa_B0030: BBa_K5299200, BBa_K5299202,BBa_K5299204.
Here you can find more information about the constructs tested containing BBa_B0034: BBa_K5299201, BBa_K5299203,BBa_K5299205.
Plated 200 ul 5 times, out of each single liquid bacterial culture, created from the same bacterial colony, in order to establish accuracy through technical repeats.
Medium used was M9 due to minimal interference, with D-glucose serving as the carbon source. Also, served as blank, plated 5 times.
Measurements were normalised as such: using the average price of fluorescence for the 5 technical repeats and dividing it by the average price of OD.
Standard deviation included in the graphs.
Results
Measurements were taken over the course of 16 hours.Here are the results for BBa_B0030 regarding BBa_J23119 with the pDGB3a1 backbone. BBa_B0030 proved less strong than BBa_B0034.

For more information regarding BBa_J23119, head over to BBa_J23119.
Here are the results for BBa_B0030 regarding BBa_J45992 in the pDGB3a1 backbone. BBa_B0030 proved less strong than BBa_B0034.

For more information regarding BBa_J45992, head over to BBa_J45992.
Here are the results for BBa_B0030 regarding P3.1 (BBa_K4583008) in the pDGB3a1 backbone. BBa_ B0030 proved less strong than BBa_B0034.

For more information regarding P3.1, head over to BBa_K4583008.
//ribosome/prokaryote/ecoli
//chassis/prokaryote/ecoli
//direction/forward
//regulation/constitutive
| biology | |
| efficiency | 0.6 |

 1 Registry Star
1 Registry Star