Bacillus subtilis
Bacillus subtilis is gram-positive model organism. Thus, much is known about this organism. The genome of Bacillus subtilis strain 168 has been sequenced.
Although the current part collection for B. subtilis is small, many are now using B. subtilis as a candidate host for synthetic devices and systems. Please read more about the advantages and disadvantages of using B. subtilis as a chassis.
| Plasmid backbones (?) | Promoters (?) | Ribosome binding sites (?) | DNA parts (?) |
Or get get help on Bacillus subtilis parts.
Plasmid backbone
| Name | Description | Resistance | Replicon | Copy number | Chassis | Length |
|---|---|---|---|---|---|---|
| BBa_I742123 | Multi-host vector pTG262 converted to BioBrick vector | C | Multi-host | Multi-host | 5564 | |
| BBa_J179000 | pBS1C: Bacillus subtilis vector, amyE integration, CAM-resistance | A(E. coli) C(B. subtilis) | B. subtilis + E. coli | 6105 | ||
| BBa_J179001 | pBS2E: Bacillus subtilis vector, lacA integration, MLS resistance | A(E. coli) E(B. subtilis) | B. subtilis + E. coli | 6258 | ||
| BBa_J179002 | pBs4S: Bacillus subtilis vector, thrC integration, spec resistance | A(E. coli) S(B. subtilis) | pDG1731 | integrative in B. subtilis | B. subtilis + E. coli | 4573 |
| BBa_K090402 | B. subtilis Episomal Vector with Constitutive GFP | 6337 | ||||
| BBa_K090403 | Gram-positive Shuttle Vector for Chromosomal Integration | 5632 | ||||
| BBa_K1065204 | Efe+Bba_B0015 in BBa_K823024 (pXyl) | A(E. coli) S(B. subtilis) | B. subtilis + E. coli | 6017 | ||
| BBa_K1085014 | pSB1AC3-HySp: Integrational backbone into B. subtilis amyE locus with IPTG inducible promoter | 7625 | ||||
| BBa_K1185004 | Integration vector for Bacillus subtilis derived from pSac-Cm | AC | multi-host | multi-host | 5210 | |
| BBa_K1364021 | Integrative plasmid for Bacillus subtilis (pSBbs4E) | 5760 | ||||
| BBa_K818000 | Integration vector for Bacillus subtilis derived from pSac-Cm | AC | Multi host | Multi host | 5223 | |
| BBa_K823021 | pSBBs1C-lacZ (lacZ reporter vector for B. subtilis) | A(E. coli) C(B. subtilis) | pAC6 | integrative in B. subtilis | E.coli and B. subtilis | 9792 |
| BBa_K823022 | pSBBs4S: Empty backbone for integration into Bacillus subtilis thrC locus | A(E. coli) S(B. subtilis) | pDG1731 | integrative in B. subtilis | B. subtilis + E. coli | 4573 |
| BBa_K823023 | pSBBs1C: Empty backbone for integration into Bacillus subtilis amyE locus | A(E. coli) C(B. subtilis) | B. subtilis + E. coli | 6105 | ||
| BBa_K823024 | pSBBs4S-Pxyl: Integrative expression vector for Bacillus subtilis | A(E. coli) S(B. subtilis) | B. subtilis + E. coli | 4794 | ||
| BBa_K823025 | pSBBs3C-luxABCDE (lux reporter vector for B.subtilis) | A(E. coli) + C(B. subtilis) | pAH328 | integrative in B. subtilis | B. subtilis + E. coli | 10640 |
| BBa_K823027 | pSBBs2E: Empty backbone for integration into Bacillus subtilis lacA locus | A(E. coli) E(B. subtilis) | B. subtilis + E. coli | 6193 |
Constitutive promoters
The promoters here are B. subtilis promoters that are constitutive meaning that the activity of these promoters should only be regulated by the levels of RNA polymerase and the appropriate σ factor.
The sequence of these promoter are adapted to the σ factor of B. subtilis. However, some of these promoter also works in E. coli. Generally speaking, standard E. coli promoters don't work (or are very weak) in B. subtilis strains, whereas the contrary generally works. However, it doesn't mean that the efficiency will be the same in both strains. The pVeg promoter, for instance, works fine at a high level of expression in both E. coli and B. subtilis strains - Contribution from User: Cyrpaut (31 October 2011)
Constitutive B. subtilis σA promoters
This section lists promoters that are recognized by B. subtilis σA RNAP. σA is the major B. subtilis sigma factor so there should be RNAP present to transcribe these promoters under most growth conditions (although maximally during exponential growth).
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_K143012 | Promoter veg a constitutive promoter for B. subtilis | . . . aaaaatgggctcgtgttgtacaataaatgt | 97 | 6561 | In stock | ||
| BBa_K143013 | Promoter 43 a constitutive promoter for B. subtilis | . . . aaaaaaagcgcgcgattatgtaaaatataa | 56 | 11873 | Not in stock | ||
| BBa_K780003 | Strong constitutive promoter for Bacillus subtilis | . . . aattgcagtaggcatgacaaaatggactca | 36 | 1559 | It's complicated | ||
| BBa_K823000 | PliaG | . . . caagcttttcctttataatagaatgaatga | 121 | 7765 | In stock | ||
| BBa_K823002 | PlepA | . . . tctaagctagtgtattttgcgtttaatagt | 157 | 7245 | In stock | ||
| BBa_K823003 | Pveg | . . . aatgggctcgtgttgtacaataaatgtagt | 237 | 14126 | In stock |
Constitutive B. subtilis σB promoters
This section lists promoters that are recognized by B. subtilis σB RNAP. σB is the major stationary phase E. coli sigma factor. Use these promoters when you want high promoter activity during stationary phase or during starvation.
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_K143010 | Promoter ctc for B. subtilis | . . . atccttatcgttatgggtattgtttgtaat | 56 | 4297 | Not in stock | ||
| BBa_K143011 | Promoter gsiB for B. subtilis | . . . taaaagaattgtgagcgggaatacaacaac | 38 | 2765 | Not in stock | ||
| BBa_K143013 | Promoter 43 a constitutive promoter for B. subtilis | . . . aaaaaaagcgcgcgattatgtaaaatataa | 56 | 11873 | Not in stock |
Positively regulated promoters
The B. subtilis promoters of this section is the ones that are said to be positively regulated. It means that meaning their expression level increase with the help of another third party protein called transcription activator (This category exclude the sigma factor protein itself). With the appropriate protein, you would be able to increase the activity of your promoter. Please read the description and characterization of each parts for more details.
Positively regulated B. subtilis σA promoters
This section lists the promoters recognized by B. subtilis σA RNA polymerase sub-unit. σA is the major B. subtilis sigma factor that is present under most growth conditions (but maximal during exponential growth phase).
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_K090504 | Gram-Positive Strong Constitutive Promoter | . . . acatgggaaaactgtatgtatttgatcctc | 239 | 2275 | It's complicated |
Positively regulated B. subtilis σB promoters
This section lists promoters that are recognized by B. subtilis σB RNA polymerase. σB is the polymerase subunit that is the most present during the stationary growth phase. You can use these promoters if you want your construct to be mostly expressed during stationary growth phase or under starvation conditions.
There are no parts for this table
Repressible promoters
The B. subtilis promoters of this section are said negativly regulated promoters, because they can be repressed by the expression of a third party protein. The inhibition can be released by the addition of a molecule, like for the LacI E. coli promoter.
In the following biobricks, the proposed promoters are build with the fusion of one or several operons with a σA type contitutive promoter.
| Name | Description | Promoter Sequence | Positive Regulators | Negative Regulators | Length | Doc | Status |
|---|---|---|---|---|---|---|---|
| BBa_K090501 | Gram-Positive IPTG-Inducible Promoter | . . . tggaattgtgagcggataacaattaagctt | 107 | 2054 | It's complicated | ||
| BBa_K143014 | Promoter Xyl for B.subtilis | . . . agtttgtttaaacaacaaactaataggtga | 82 | 2112 | Not in stock | ||
| BBa_K143015 | Promoter hyper-spank for B. subtilis | . . . aatgtgtgtaattgtgagcggataacaatt | 101 | 2527 | Not in stock |
In the future, we may also find promoters builded with the σB promoter.
There are no parts for this table
Ribosome binding sites
| Name | Sequence | Description | Relative Strength | Predicted Strength |
|---|---|---|---|---|
| BBa_K090505 | aaaggaggtgt | ''Bacillus subtilis'' consensus RBS | ||
| BBa_K090506 | agaggtggtgt | ''Bacillus subtilis'' weak RBS | ||
| BBa_K104003 | agggggccg | RBS_spaR | ||
| BBa_K104005 | ggggcgttg | RBS_spaK | ||
| BBa_K104009 | agaggagg | RBS_gfp | ||
| BBa_K1351028 | aaggagggata | LMU Bacillus RBS collection 1 | ||
| BBa_K1351029 | agaggaggata | LMU Bacillus RBS collection 2 | ||
| BBa_K1351030 | aaggagagata | LMU Bacillus RBS collection 3 | ||
| BBa_K1351031 | aggagaggata | LMU Bacillus RBS collection 4 | ||
| BBa_K1351032 | aagaggagata | LMU Bacillus RBS collection 5 | ||
| BBa_K1351033 | agaaagggata | LMU Bacillus RBS collection 6 | ||
| BBa_K1351034 | aagaagagata | LMU Bacillus RBS collection 7 | ||
| BBa_K143020 | taaaggaggaa | GsiB ribosome binding site (RBS) for B. subtilis | ||
| BBa_K143021 | aaaggtggtgaa | SpoVG ribosome binding site (RBS) for B. subtilis | ||
| BBa_K2586010 | . . . tgctacaattactagaggtttaaaggagga | Palf4-RBS: Palf4 linked to a RBS | ||
| BBa_K3831005 | gtaataagtaggttaggagagg | Synthetic RBS_a | ||
| BBa_K3831006 | . . . actacacgacaattaaagaaggtatttttt | Synthetic RBS_b | ||
| BBa_K3831007 | ggtggaaaggaggtgatcgac | Native RBS R2 | ||
| BBa_K3831008 | ggtgggaaggagggggttcgac | RBS R6 | ||
| BBa_K3831009 | gggatagacccagggggaggttttttt | Synthetic RBS_c | ||
| BBa_K3831014 | gctcttaaggaggattttaga | Native RBS R1 | ||
| BBa_K3831018 | gattaactaataaggaggacaaac | Native RBS R0 | ||
| BBa_K3831020 | ggtgggaaggaggacattcgac | Artificial RBS R3 | ||
| BBa_K3831047 | aaaaaaacctccccctgggtctatccc | Synthetic RBS_c - reversed | ||
| BBa_K3831049 | gtcgaaccccctccttcccacc | RBS R6 - reversed | ||
| BBa_K3831051 | gtcgatcacctcctttccacc | RBS R2 - reversed | ||
| BBa_K3831053 | . . . aaataccttctttaattgtcgtgtagtaat | Synthetic RBS_b - reversed | ||
| BBa_K3831055 | cctctcctaacctacttattac | Synthetic RBS_a - reversed | ||
| BBa_K4382009 | aaggaggaaggatca | RBS for Bacillus Subtilis | ||
| BBa_K780001 | atattaagaggaggag | Strong RBS for Bacillus Subtilis | ||
| BBa_K780002 | agagaacaaggagggg | Strong RBS for Bacillus Subtilis |
DNA parts
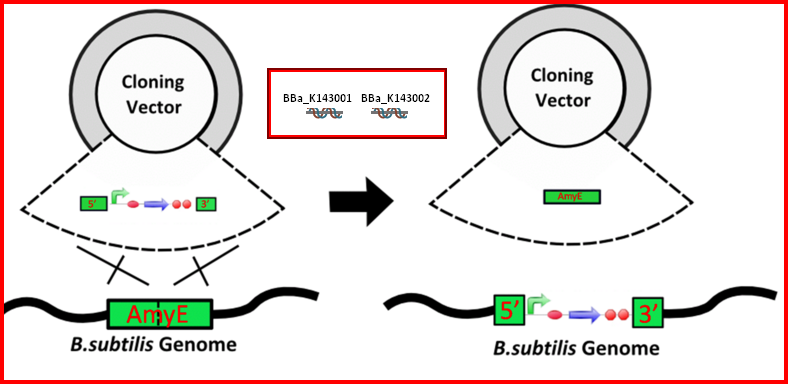
| The AmyE integration DNA parts (BBa_K143001 and BBa_K143002) are two parts that can be added to the 5' and 3' ends of a construct to allow integration into the B. subtilis genome. These parts have been successfully used within the parts BBa_K143079 and BBa_K143082 for integration. Integrated synthetic biological systems offer better genetic stability and more regulated copy number than plasmid-borne systems. For more information about these parts, please see [http://2008.igem.org/Team:Imperial_College 2008 Imperial College iGEM team wiki.] |
| Name | Description | Sequence | Length |
|---|---|---|---|
| BBa_K143001 | 5ĺ Integration Sequence for the amyE locus of B. subtilis | . . . aacacacaaattaaaaactggtctgatcga | 522 |
| BBa_K143002 | 3ĺ Integration Sequence for the amyE locus of B. subtilis | . . . tcgggcttaagcggttctcttccccattga | 1002 |
| BBa_K3831010 | Spacer_0 | . . . cttactctgttgaaaacgaatagataggtt | 40 |
| BBa_K3831011 | Spacer_1 | tgctcgtagtttacc | 15 |
| BBa_K3831012 | Spacer_5 | . . . agaatagtcaatcttcggaaatcccaggtg | 40 |
| BBa_K3831015 | Spacer_7 | taataaaaggtcccg | 15 |
| BBa_K3831023 | Spacer 03 | aaggaacggttattt | 15 |
| BBa_K3831026 | Spacer 02 | agattactactgata | 15 |
| BBa_K3831027 | Spacer 06 | ccgattctgagacgg | 15 |
| BBa_K3831028 | Spacer 04 | . . . aatacaggacccgaatcgtttcagttgcct | 40 |
| BBa_K3831038 | Spacer_SP1 | . . . ttaccacggatacagacagtgataatctta | 40 |
References
Given the number of available articles on B. subtilis, we only include some review articles here.
<biblio>
- Earl pmid=18467096
- Pavlendova pmid=18450217
- Sonenshein pmid=17982469
- Lopez pmid=17981078
- Aguilar pmid=17977783
- Irnov pmid=17381303
</biblio>