Phytobrick version of 5'Connector Dummy
This is the Phytobrick version of the Connector 5'Connector Dummy and was build as a part of the Marburg Collection. Instructions of how to use the Marburg Collection are provided at the bottom of the page.
Overview
One novel key feature of our toolbox are the connectors. They were designed in order to function as insulators to prevent crosstalk between neighboring transcription units (Design of the Marburg Collection). Therefore a perfectly insulating connector would prevent the readthrough from backbone sequences that most probably caused the notably high expression that was measured in the promoter experiment for the dummy promoter (Promoter experiment). In addition to blocking transcriptional readthrough, a good connector must not possess any cryptic promoter activity.
In already existing toolboxes, the combinatorics that come with choosing oris and resistances would be multiplied by the number of required positional vectors which provide the fusion sites for LVL2 assembly. A toolbox with five possible positions, four resistances and three oris would require 60 LVL1 destination plasmids. To enable inversion of transcription units, a built-in feature in our toolbox, the previous number would be multiplied by two, resulting in 120 required LVL1 destination plasmids. Unsurprisingly, this theoretical toolbox would not be convenient to be built and used.
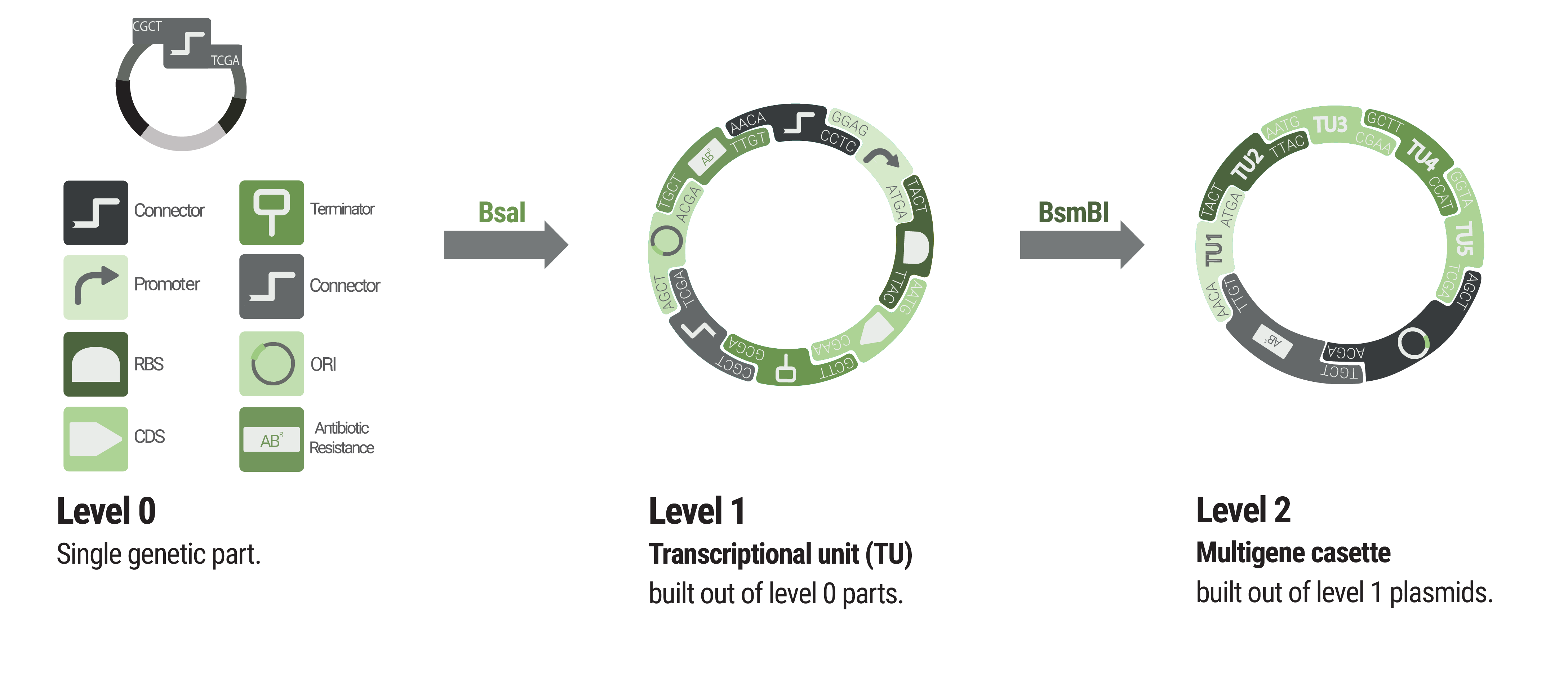
The Marburg Collection presents a novel approach that achieves maximum flexibility with a minimum of required components (figure 1).
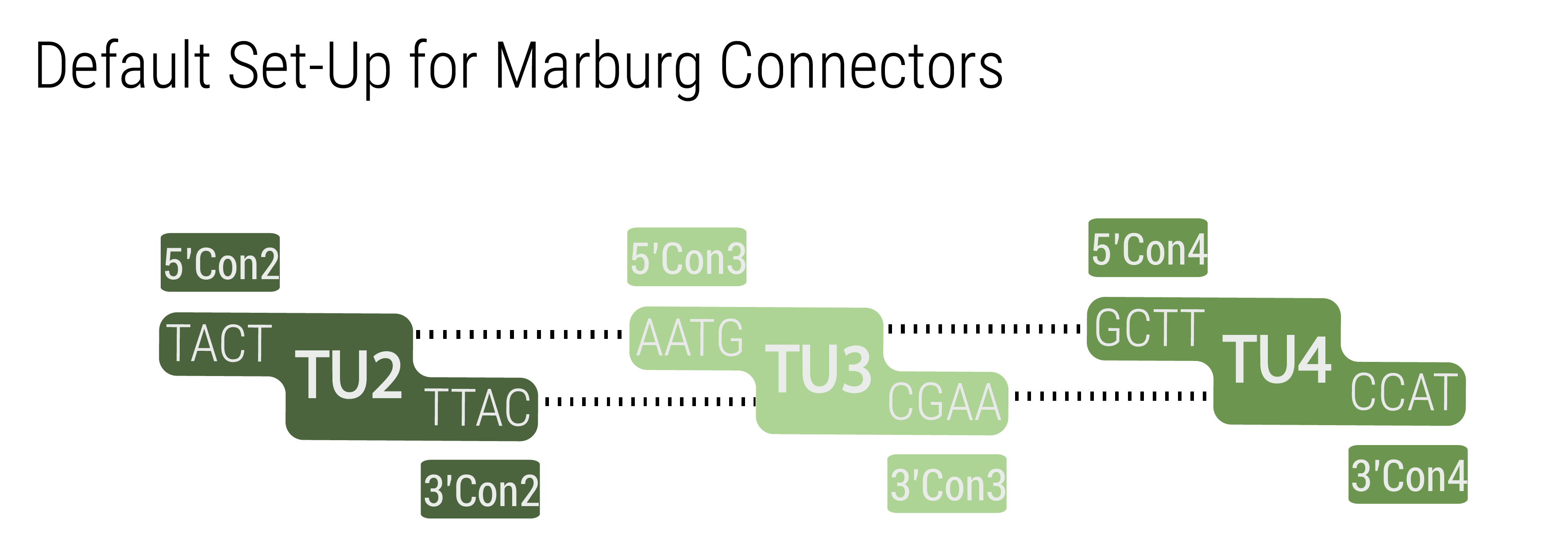
We provide two sets of connectors. The first set, the “short connectors” solely provide the fusion sites for subsequent assembly. The fusion sites are identical to the ones that are used in building LVL1 plasmids to avoid having to design a complete new set of fusion sites. This also enables using LVL0 ori and resistance parts in LVL2 plasmids. Inversion of individual transcription units was also achieved by designing fusion sites that match in reverse order.
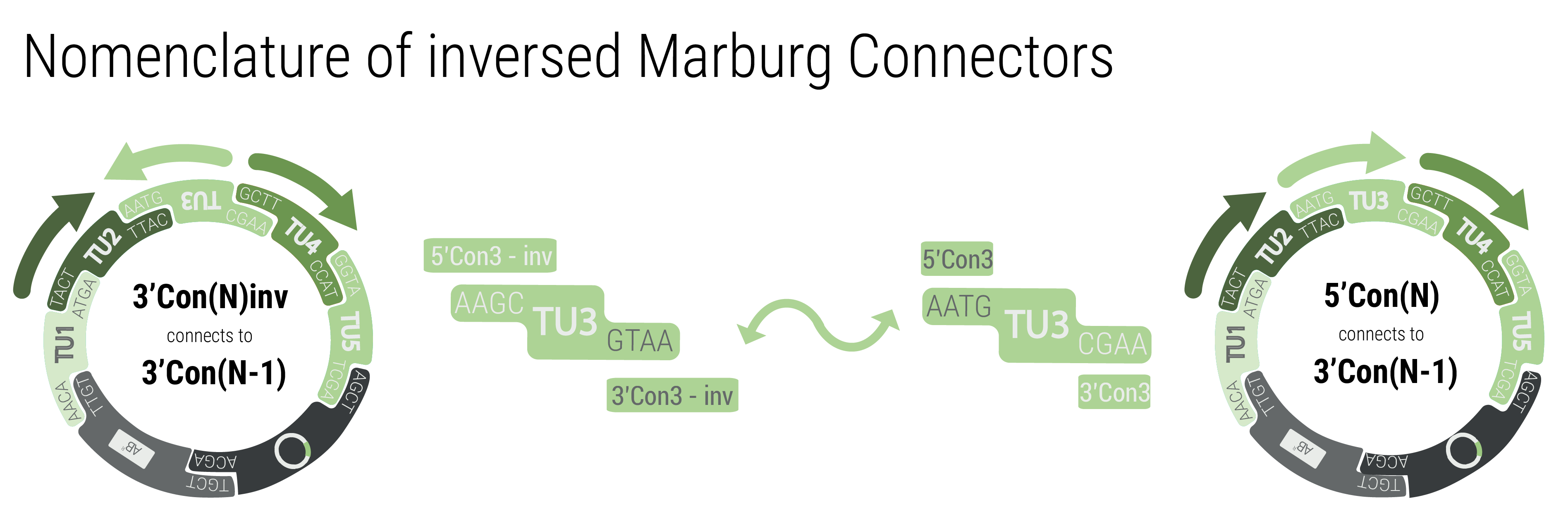
To achieve inversion, the respective transcription unit has to be built with connectors that match the neighboring transcription units. The function by following pattern: 3'Con(N)inv connects to 3'Con(N-1) and 5'Con(N)inv connects to 5'Con(N+1)
The 5’ and 3’ connectors can be chosen independently to build a LVL1 plasmid. This allows the user to combine any number of LVL1 plasmids between one and five into a LVL2 plasmid without end linkers that are required in other toolboxes (Andreou and Nakayama, 2018). An unusual example is shown in figure xxx to demonstrate the number of possibilities. The only logic that has to be followed for designing multigene constructs is to follow the formula 5'Con(N) connects to 3'Con(N-1)<b>. In the default set up TU(N) is built with 5'Con(N) and 3'Con(N) This will result in a LVL2 plasmid harboring five transcription units. However, any other combination that follows the previously given rule will result in a functionally assembled LVL2 plasmid.
Characterization
We focused on characterizing the 5’ Connector because we expect the stronger influence on signal strengths. For characterizing our connector parts, we created 20 test plasmids with the lux operon as the reporter.
In our toolbox we provide five short connectors, which solely possess the fusion sites for LVL2 cloning, and five long connectors which additionally harbor self-designed insulators. Each of these ten connectors were cloned with the constitutive promoter K2560007 (J23100), to check for effects on an active promoter, and with the Promoter Dummy to quantify the extent of transcriptional activity that reaches the Promoter Dummy.
 A
A B
B
Results of Connector measurmenet
A) Connector constructs built with J23100 as promoter part
B) Connector constructs built with the Dummy Promoter as promoter part
Result
The acquired data are shown in figure 1. The data were normalized over the test construct J23100, that was used in the promoter experiment and constructed with the connector dummies. For the five constructs with the active promoter and the long connectors we observed extremely varying signals. We measured a range from 0.2 to 2 fold change compared to the reference construct. It has been shown that the sequence directly upstream of small synthetic promoters can greatly impact the transcription efficiency ( Carr et al.2017). In case of the long connectors, the sequence upstream of the promoter forms the terminator and could affect the efficiency of RNA-polymerase binding to the -35 and -10 regions. For the constructs built with small connectors, we also observed varying signals but to a lesser extent compared to the long connectors.
For all ten connectors that are provided in our toolbox, we show a tenfold range in the measured luminescence/OD600 signal. As a conclusion, we recommend to carefully consider the combination of promoter and 5’ Connector for rationally designing constructs.
Taking a look at the constructs that were built with the Promoter Dummy, we also see a huge difference in the expression signals. For the long connectors we expected a negligibly low reporter expression which we observed for two out of five long 5’ Connectors resulting in a 14 fold signal reduction compared to the “Promoter Dummy” reference. The remarkably strong signal observed for the remaining three connectors could be due to inefficient terminators or cryptic promoters in the pretended “neutral sequence”.
For the remaining five constructs possessing the five short 5’ connectors we observed a range from 0.3 to 5.5 fold compared to the “Promoter Dummy” reference. We are not able to give an experimental explanation for this observation but we could imagine that the LVL2 fusion sites, the only four bases that differ in these constructs, could constitute a weak promoter together with surrounding sequences.
Summarizing the connector characterization, we found that sequences upstream of short synthetic promoters greatly affect reporter expression, which is in accordance with literature ( Carr et al.2017). Moreover, we demonstrated that two of our five self-designed connectors efficiently reduce the signal resulting from other sources than the actual promoter.
We additionally conclude that algorithms that predict the “neutrality” of sequences alone are not sufficient to create well functioning insulators.