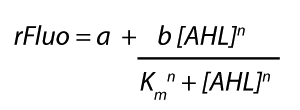Difference between revisions of "Part:BBa C0062:Experience"
(→Results) |
(→Results) |
||
| Line 76: | Line 76: | ||
|+'''Table 1''' Crosstalk matrix for the regulator LuxR ([https://parts.igem.org/Part:BBa_C0062:Experience BBa_C0062]) | |+'''Table 1''' Crosstalk matrix for the regulator LuxR ([https://parts.igem.org/Part:BBa_C0062:Experience BBa_C0062]) | ||
|colspan="4" style='font-size:10pt';text-align:left| | |colspan="4" style='font-size:10pt';text-align:left| | ||
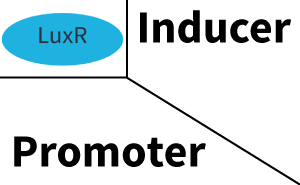
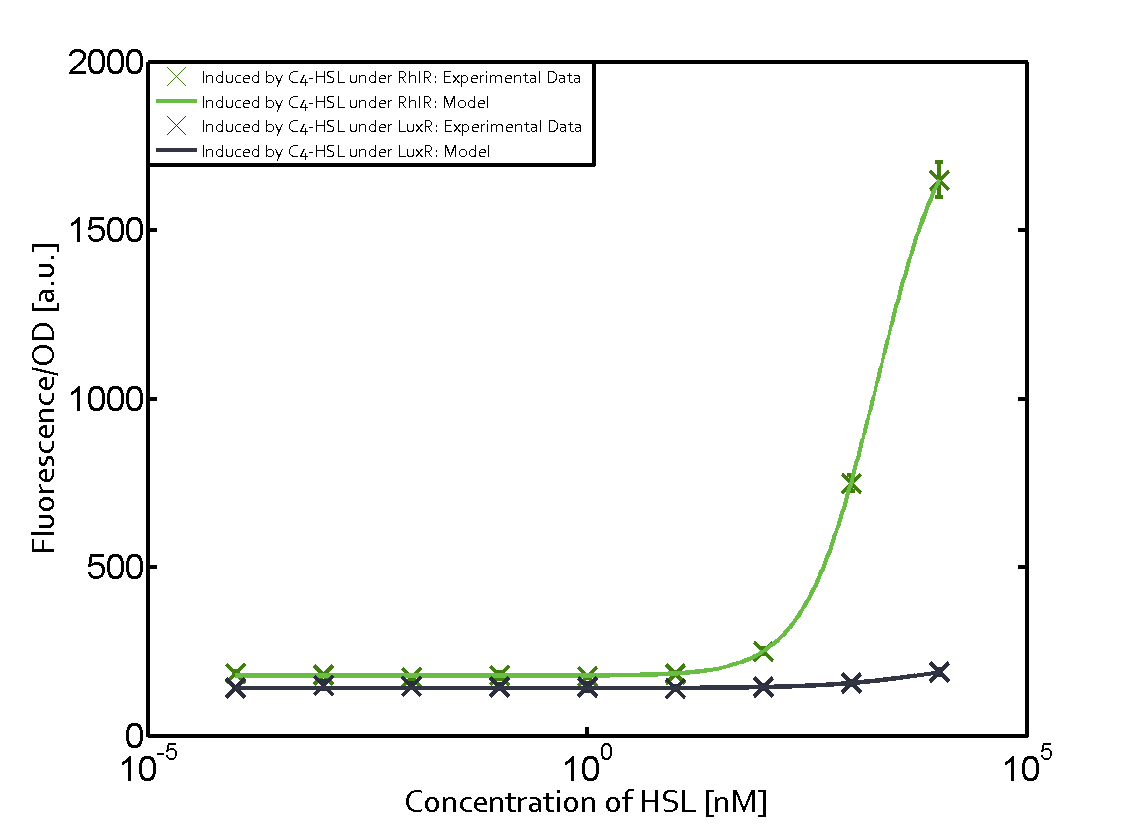
| − | In all the measurements conducted to create this matrix the [https://parts.igem.org/Part:BBa_C0062 regulator LuxR] was the basis and was induced in six different variations shown. Colored curves indicate the coventional inducer/regulator/promoter combination, black curves indicate crosstalk. | + | In all the measurements conducted to create this matrix the [https://parts.igem.org/Part:BBa_C0062 regulator LuxR] was the basis and was induced in six different variations shown. Colored curves indicate the coventional inducer/regulator/promoter combination (e.g. [[3OC6HSL|3OC6-HSL]]/[[Part:BBa_C0062|LuxR]]/[[Part:BBa_R0062|pLux]]), black curves indicate crosstalk. |
|- | |- | ||
|style="width:200px"|[[File:ETH_Zurich_2014_qs-table_Corner_LuxR.png|100px|link=https://parts.igem.org/Part:BBa_C0062:Experience]] | |style="width:200px"|[[File:ETH_Zurich_2014_qs-table_Corner_LuxR.png|100px|link=https://parts.igem.org/Part:BBa_C0062:Experience]] | ||
Revision as of 10:31, 28 October 2014
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_C0062
User Reviews
UNIQ870aa20be2899e34-partinfo-00000000-QINU
|
••••
ETH Zurich 2014 |
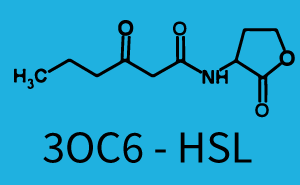
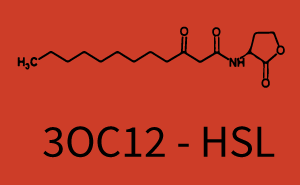
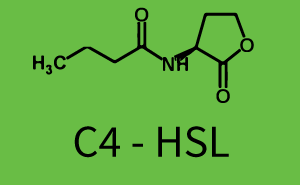
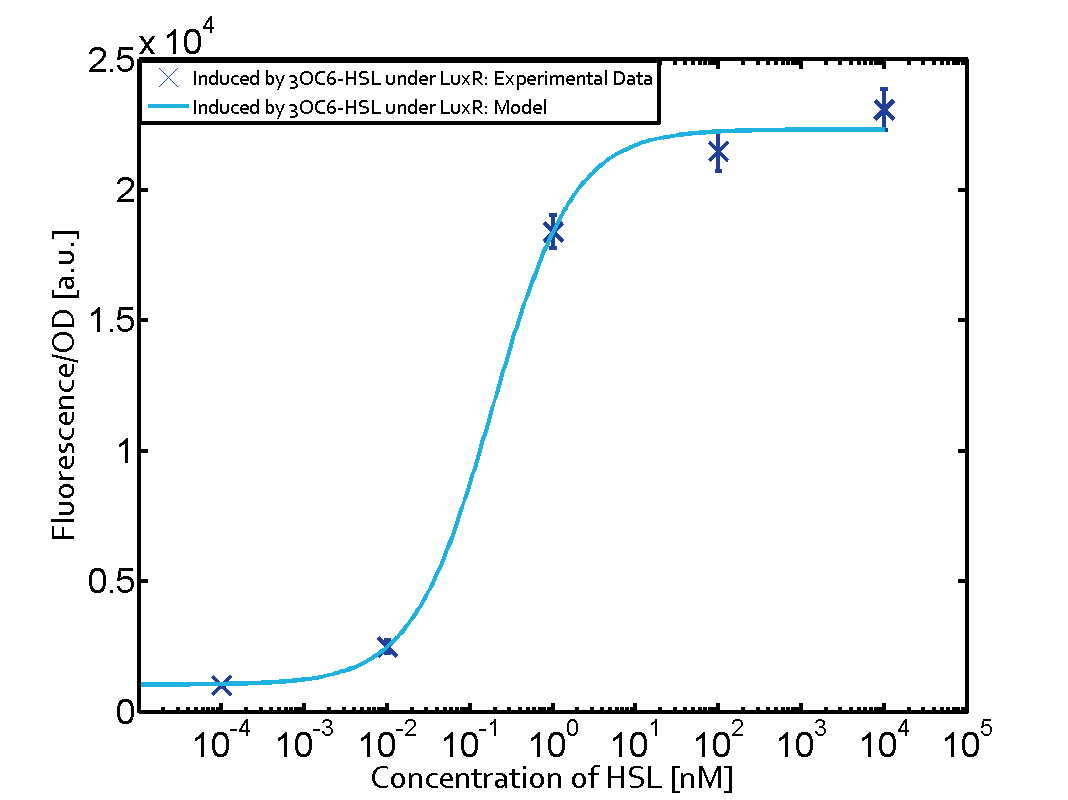
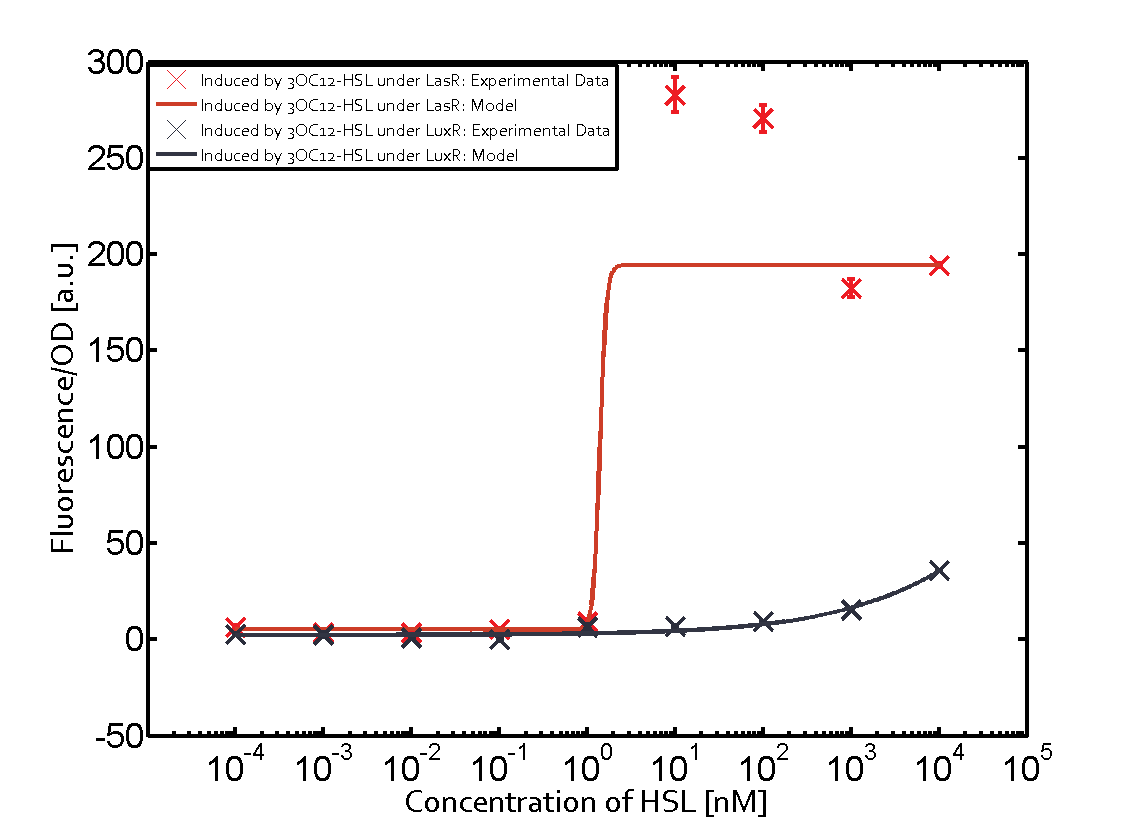
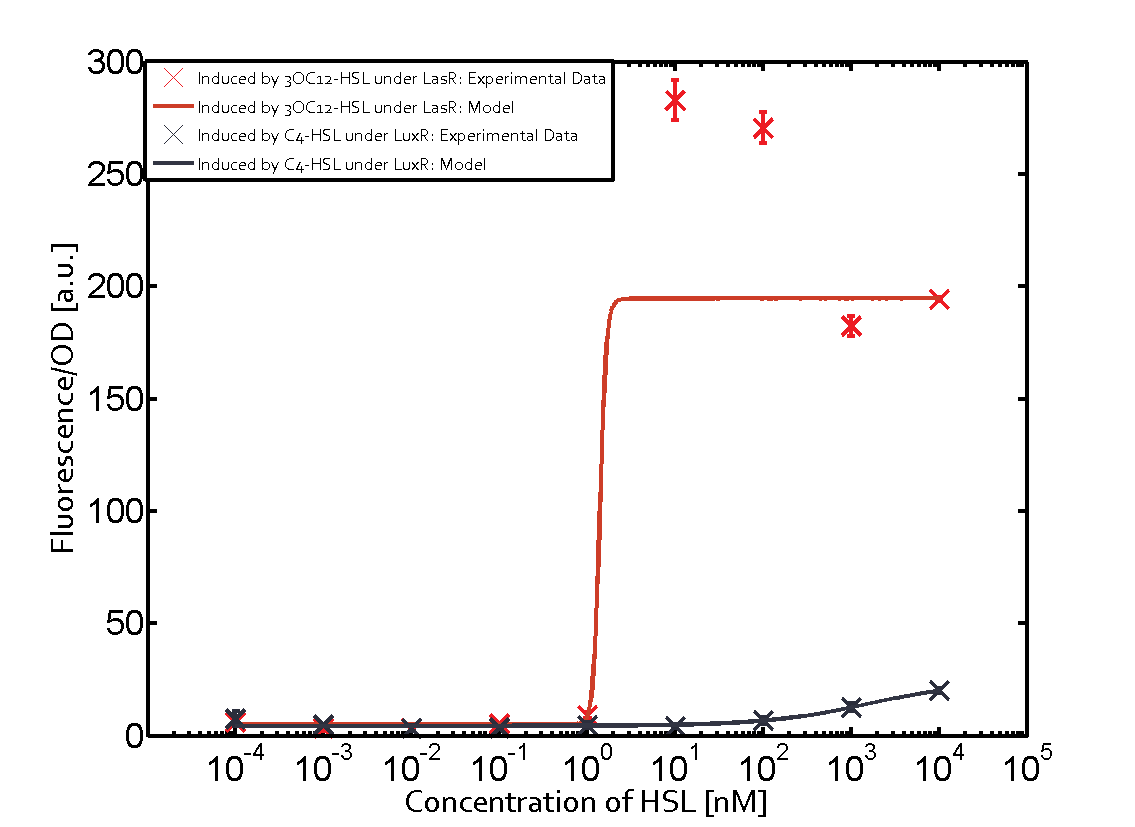
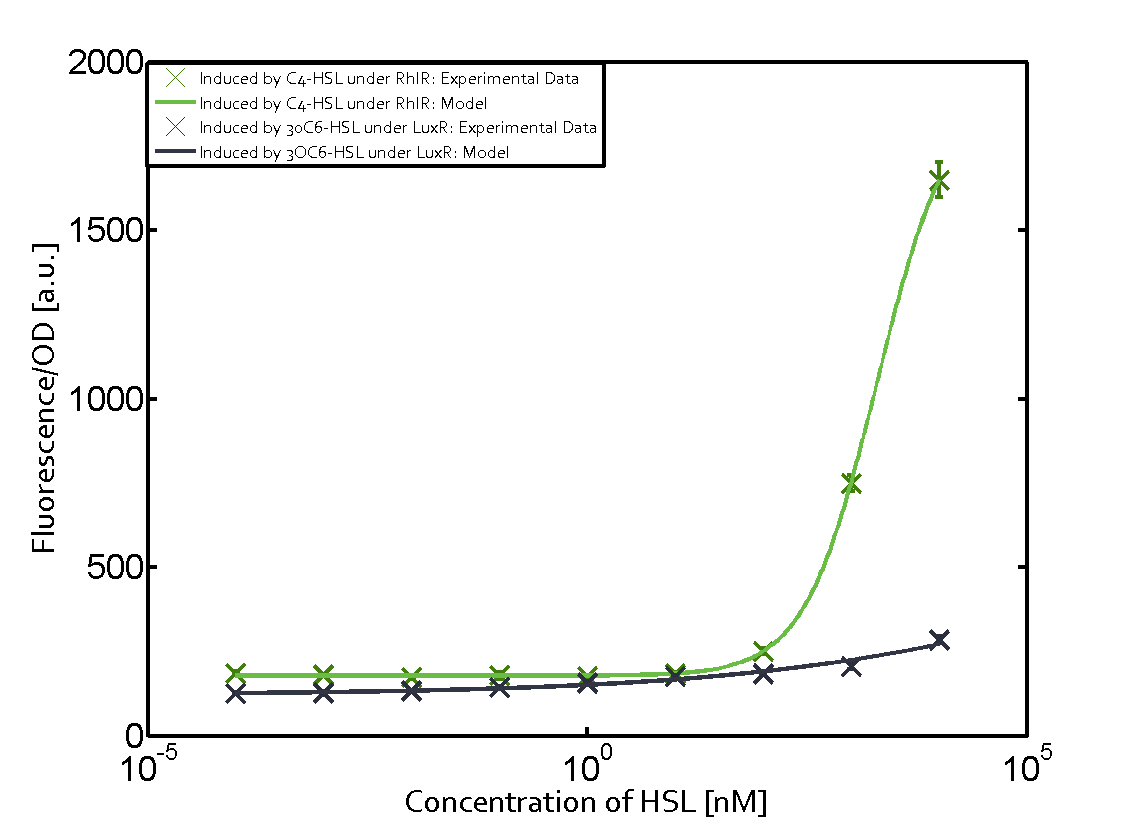
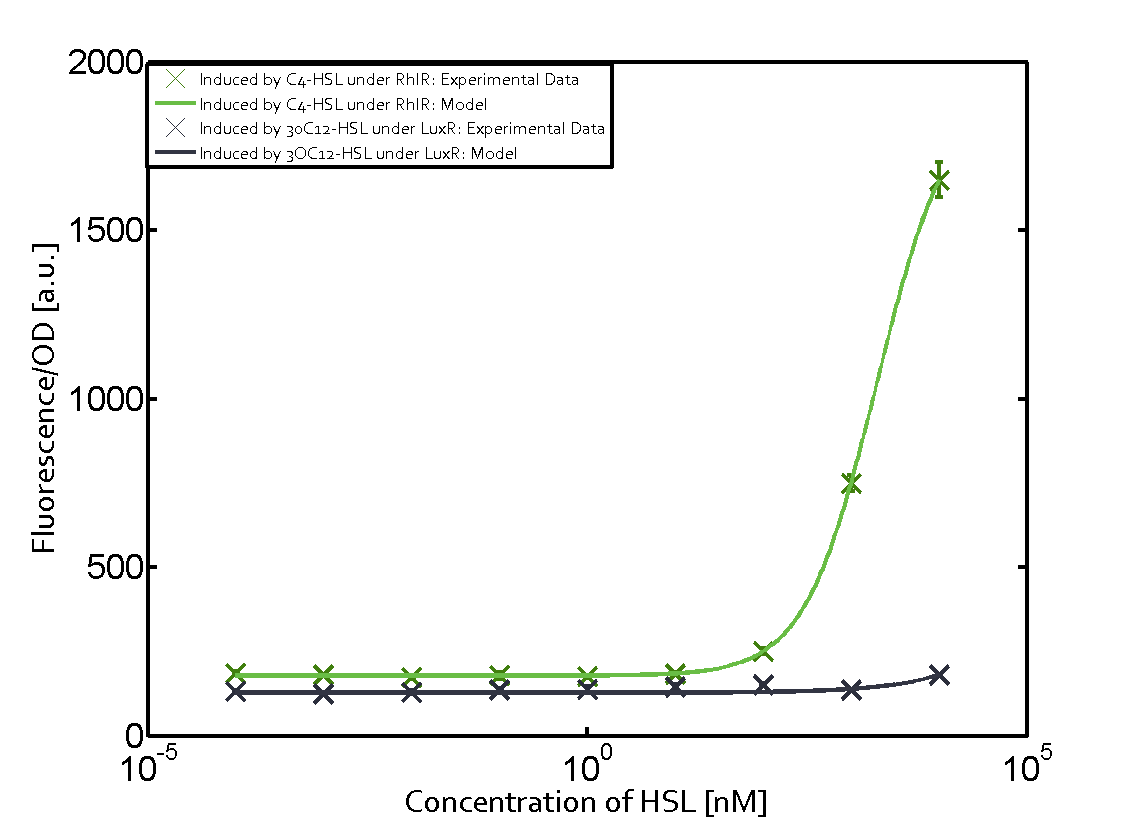
Background informationWe used an E. coli TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and promoter pLux (BBa_R0062) followed by RBS (BBa_B0034) and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts BBa_B0040 and BBa_B0015 were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, and one of three promoters chosen from the Anderson promoter collection followed by luxR (BBa_C0062). The detailed regulator construct design and full sequences (piG0041, piG0046, piG0047) are [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. Experimental Set-UpThe above described E. coli TOP10 strains were grown overnight in Lysogeny Broth (LB) containing kanamycin (50 μg/mL) and ampicillin (200 μg/mL) to an OD600 of about 1.5 (37 °C, 220 rpm). As a reference, a preculture of the same strain lacking the sfGFP gene was included for each assay. The cultures were then diluted 1:40 in fresh LB containing the appropriate antibiotics and measured in triplicates in microtiter plate format on 96-well plates (200 μL culture volume) for 10 h at 37 °C with a Tecan infinite M200 PRO plate reader (optical density measured at 600 nm; fluorescence with an excitation wavelength of 488 nm and an emission wavelength of 530 nm). After 200 min we added the following concentrations of inducers (3OC6-HSL, 3OC12-HSL, and C4-HSL): 10-4 nM and 104 nM (from 100 mM stocks in DMSO). Attention: All the dilutions of 3OC12-HSL should be made in DMSO in order to avoid precipitation. In addition, in one triplicate only H2O was added as a control. From the the obtained kinetic data, we calculated mean values and plotted the dose-response-curves for 200 min past induction. Characterization of the promoter's sensitivity to 3OC6-HSL depending on LuxR concentrationThe amount of regulator LuxR (BBa_C0062) in the system was shown to influence the pLux promoter's response to the inducer concentration (3OC6-HSL). By using the three different constitutive promoters BBa_J23100, BBa_J23109, and BBa_J23111 for the production of LuxR we have measured this effect in terms of fluorescence (see Figure 1). ResultsThe measurements of the induced system with 3OC6-HSL concentrations of 10-13 M to 10-5 M showed an increasing sensitivity of the pLux (BBa_R0062) promoter (in terms of fluorescence per OD600) for increasing strength of the promoter controlling LuxR (BBa_C0062) expression. For BBa_J23100 (strongest promoter chosen) the sensitivity is highest (half maximal effective concentration EC50 approximately 20 pM), for BBa_J23109 (weakest one chosen) the sensitivity is lowest (EC50 approximately 100 pM), with BBa_J23111 (medium) falling between these two but closer to the strong promoter (EC50 approximately 10 nM). Overall, this is in line with the promoter strength given in the Anderson collection and suggests that the sensitivity of the pLux (BBa_R0062) promoter increases with an increase of LuxR (BBa_C0062) expression.
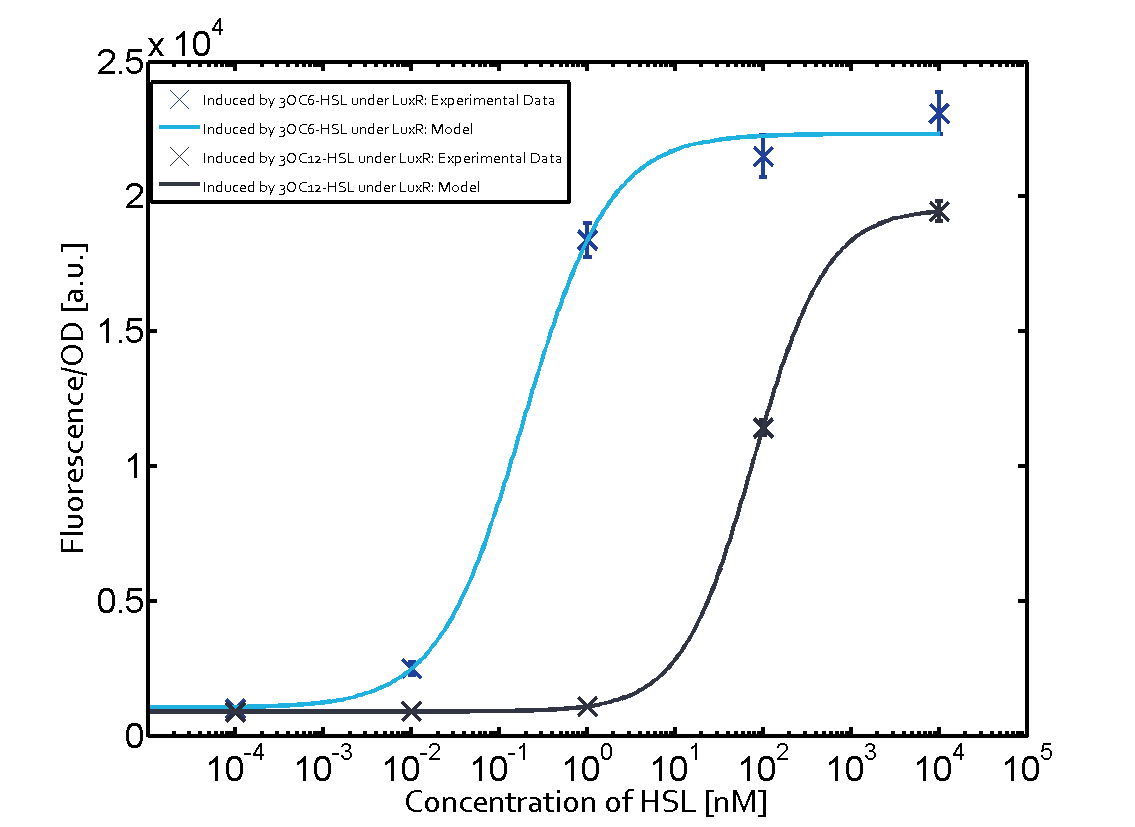
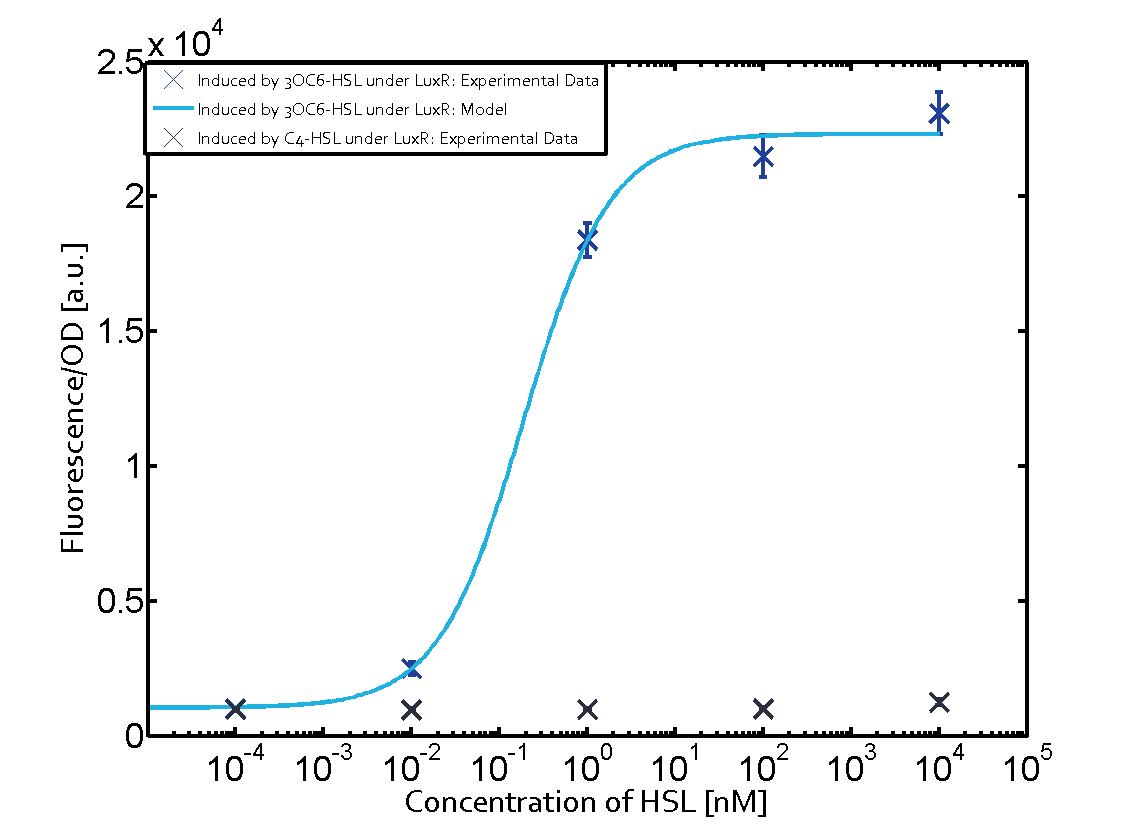
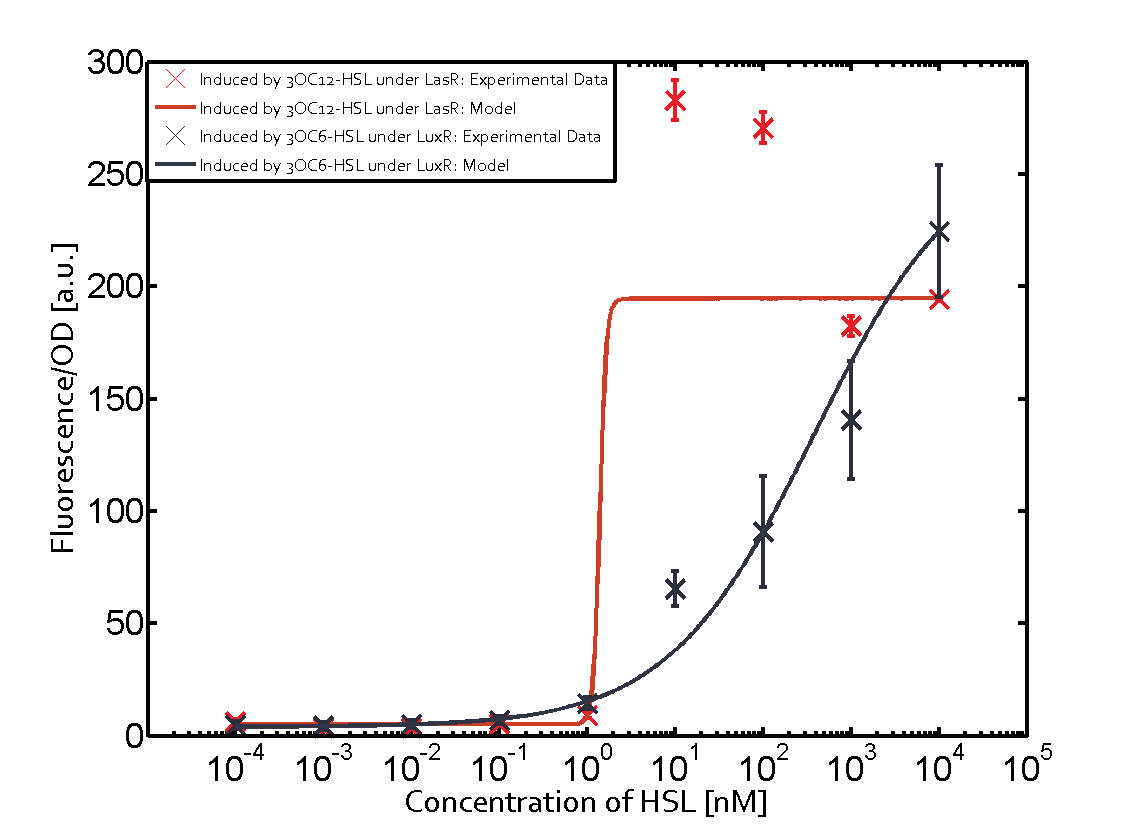
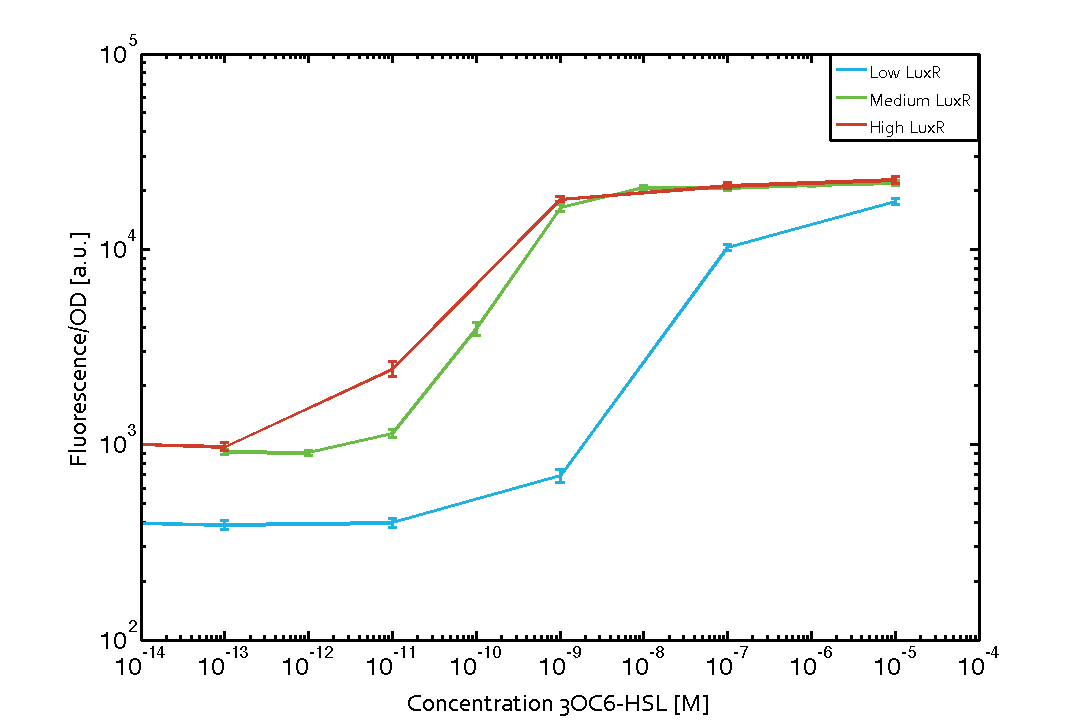 Figure 1 Effect of varied promoter strength for the expression of LuxR (BBa_C0062) and the resulting change of pLux (BBa_R0062) sensitivity. The fluorescence per OD600 is shown over an inducer-range of 10-13 M to 10-5 M. The promoters used are: BBa_J23100 (high LuxR expression, red), BBa_J23111 (medium LuxR expression, green), and BBa_J23109 (low LuxR expression, blue). All three promoters are part of the Anderson collection. Data points are mean values of triplicate measurements in 96-well microtiter plates 200 min after induction ± standard deviation. For the full data set and kinetics please [http://2014.igem.org/Team:ETH_Zurich/contact contact] us or visit the [http://2014.igem.org/Team:ETH_Zurich/data/raw raw data] page. Characterization of crosstalkBackground informationHere, we focus on the characterization of crosstalk of LuxR with different AHLs and further crosstalk of LuxR-3OC6-HSL with the three promoters - pLux, pLas, and pRhl. In the following, we describe all the different levels of crosstalk we have assessed. First-order crosstalkIn the first order crosstalk section we describe activation of pLux due to LuxR binding to inducers different from 3OC6-HSL or pLux itself binding a regulator-inducer pair different from LuxR-3OC6-HSL. First Level crosstalk: LuxR binds to different AHLs and activates the promoter pLuxIn the conventional system 3OC6-HSL binds to its corresponding regulator, LuxR, and activates the pLux promoter (Figure 2, light blue). However, LuxR can potentially also bind to other AHLs and then activate pLux (Figure 2, 3OC12-HSL in red and C4-HSL in green). 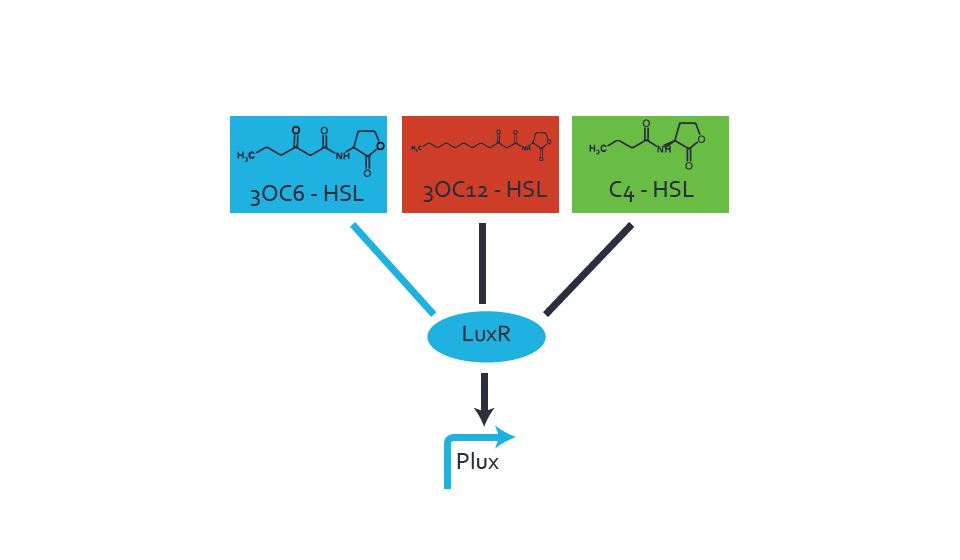 Figure 2 Overview of possible crosstalk of the LuxR/pLux system with three different AHLs. Usually, 3OC6-HSL binds to its corresponding regulator, LuxR, and activates the pLux promoter (light blue). However, LuxR may also bind 3OC12-HSL (red) or C4-HSL (green) and then unintentionally activate pLux. Second Level crosstalk: LuxR binds to 3OC6-HSL, its natural AHL, and activates promoters different from pLuxIn the conventional system 3OC6-HSL binds to its corresponding regulator, LuxR, and activates the pLux promoter (Figure 3 left-hand side, light blue). However, LuxR can potentially also bind to additional cell-cell signaling promoters (Figure 2, pLas in red and pRhl in green) and then activate genes different from the pLux regulated gene of interest . 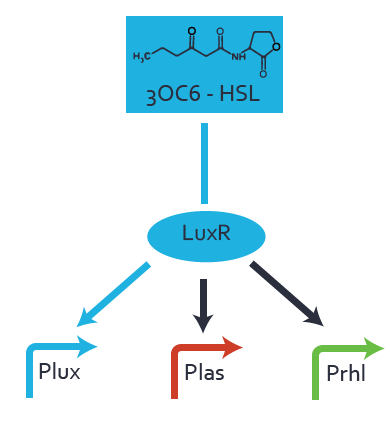 Figure 3 Overview of possible crosstalk of the regulator LuxR with three different cell-cell signaling promoters. Usually, 3OC6-HSL binds to its corresponding regulator, LuxR, and activates the pLux promoter (light blue). However, LuxR may also bind pLas (red) or pRhl (green) and then unintentionally activate 'off-target' gene expression. Second order crosstalk: Combination of both cross-talk levelsThe second order crosstalk describes unintended activation of a cell-cell signaling promoter by a mixture of both the levels described above. The regulator LuxR binds an inducer different from 3OC6-HSL and then activates a promoter different from pLux. For example, the inducer C4-HSL (green), usually binding to the regulator RhlR, could potentially interact with the LuxR regulator (light blue) and together activate the promoter pLas (red). This kind of crosstalk is explained in Figure 3. 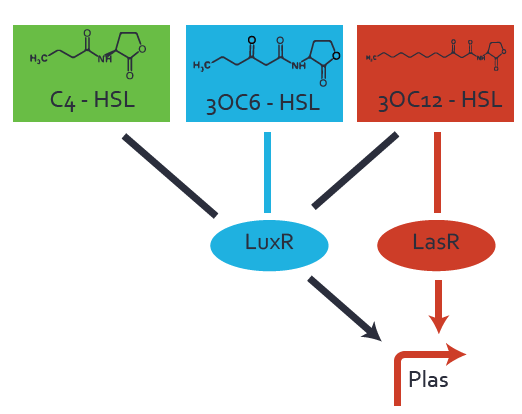 Figure 3 Overview of possible crosstalk of the pLas promoter with both the regulator and inducer being unrelated to the promoter and each other. Usually, LasR together with inducer 3OC12-HSL activate their corresponding promoter pLas (red). However, pLas may also be activated by another regulator together with an unrelated inducer. For example, the inducer C4-HSL (green) may interact with the LuxR regulator (light blue) and together activate pLas (red). Results
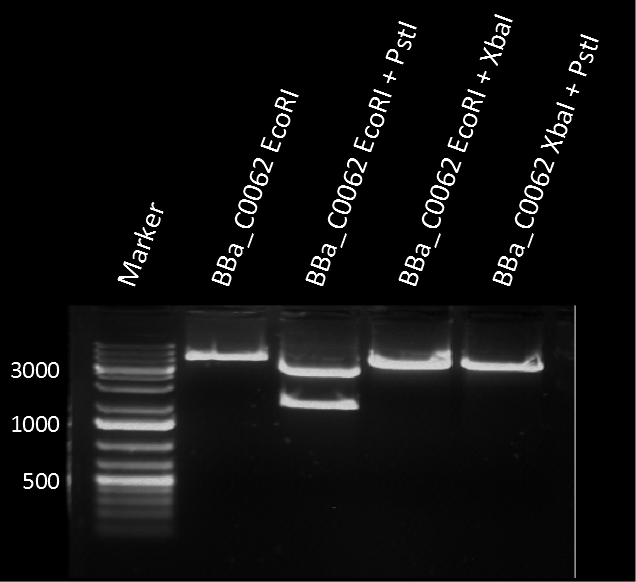
Modeling crosstalkEach experimental data set was fitted to an Hill function using the Least Absolute Residual method. The fitting of the graphs was performed using the following equation :
| ||||||||||||||||||||||||||||||||||||
|
•••••
SUN(Tsinghua) |
Part was sequenced and functional. LuxR was used in our Portable Pathogen Detector. |
|
•••••
wmholtz |
Using this part, I have successfully constructed and tested a quorum sensing circuit in E. coli. |
|
•••••
Youri |
This part was used and tested as a subpart in K546000, K546001, K546002, K546003, K546005 and K546546. This part functioned in all cases. |
|
Kevin (iGEM Braunschweig 2013) |
The plasmid pSB1C3 BBa_C0062 from the 2013 distribution Kit was transformed in E. coli XL1 BlueMRF. Sequencing with standard verification primer VF2 confirmed matching sequence of backbone DNA up to the EcoRI restriction site. The rest of the sequence (not shown) does not match the registry entry.
96 145
pSB1C3 LuxR (96) GAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATT
C0062 VF2 (1) GAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATT
146 195
pSB1C3 LuxR (146) TCTGGAATTCGCGGCCGCTTCTAGAGATGAAAAACATAAATGCCGACGAC
C0062 VF2 (51) TCTGGAATTCGACGCAA-TGGGTGCGCTGTCTACTAAATACAACGACACC
196 245
pSB1C3 LuxR (196) ACATACAGAATAATTAATAAAATTAAAGCTTGTAGAAGCAATAATGATAT
C0062 VF2 (100) CCGGAAAAAGCCTCCCGTACTTACGACGCTCACCGTGACGGTTTCGTTAT
246
pSB1C3 LuxR (246) TAATCAATGC...
C0062 VF2 (150) CGCTGGCGGC...
A restriction assay (Figure 1) showed that the sequenced part has no XbaI restriction site following the EcoRI site indicating another part in front of BBa_C0062 with a length of at least 1000 bp. |
UNIQ870aa20be2899e34-partinfo-00000007-QINU