Part:BBa_C0179
lasR activator from P. aeruginosa PAO1(no LVA)
same as C0079 except no LVA tag
New Background Information of LasR by WHU-China 2020
Group: WHU-China 2020
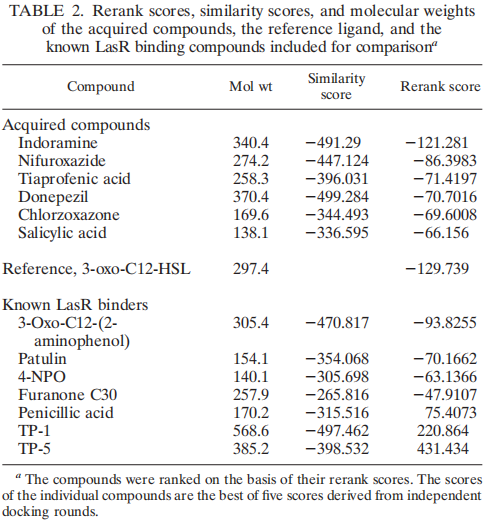
In our project this year, LasR is not only a component in the quorum sensing genetic circuit, but a target protein in the pathogenic bacteria P. aeruginosa for virulence inhibition [1]. Thus we collected information of LasR structure and LasR inhibitors, which is shown in the references [2,3]. Also, the workflow combining in silico prediction with in vivo examination sets a good example for iGEM teams.
Reference: 1. Grandclement C et al., Quorum quenching: role in nature and applied developments. FEMS Microbiology Reviews, 40 (2016): 86-116.
2. Yang L et al., Computer-Aided Identifification of Recognized Drugs as Pseudomonas aeruginosa Quorum-Sensing Inhibitors. Antimicrobial Agents and Chemotherapy, 53 (2009): 2432–2443.
3. https://www.rcsb.org/structure/2UV0
Process measurement characterisation of LasR by Shanghaitech iGEM 2017
Group: Shanghaitech 2017
We improved this part by characterising the crosstalk between LasR and non-cognate AHLs. Comparing with ‘Imperial College London iGEM 2016’ team, this year we have done two more experiments for improving this part:
- 1) We test crosstalk with two new kinds of AHL molecules – Tra 3OC8-HSL and Rpa Coumaroyl-HSL.
- 2) Our fluorescence measurement is real-time process rather than a time point.
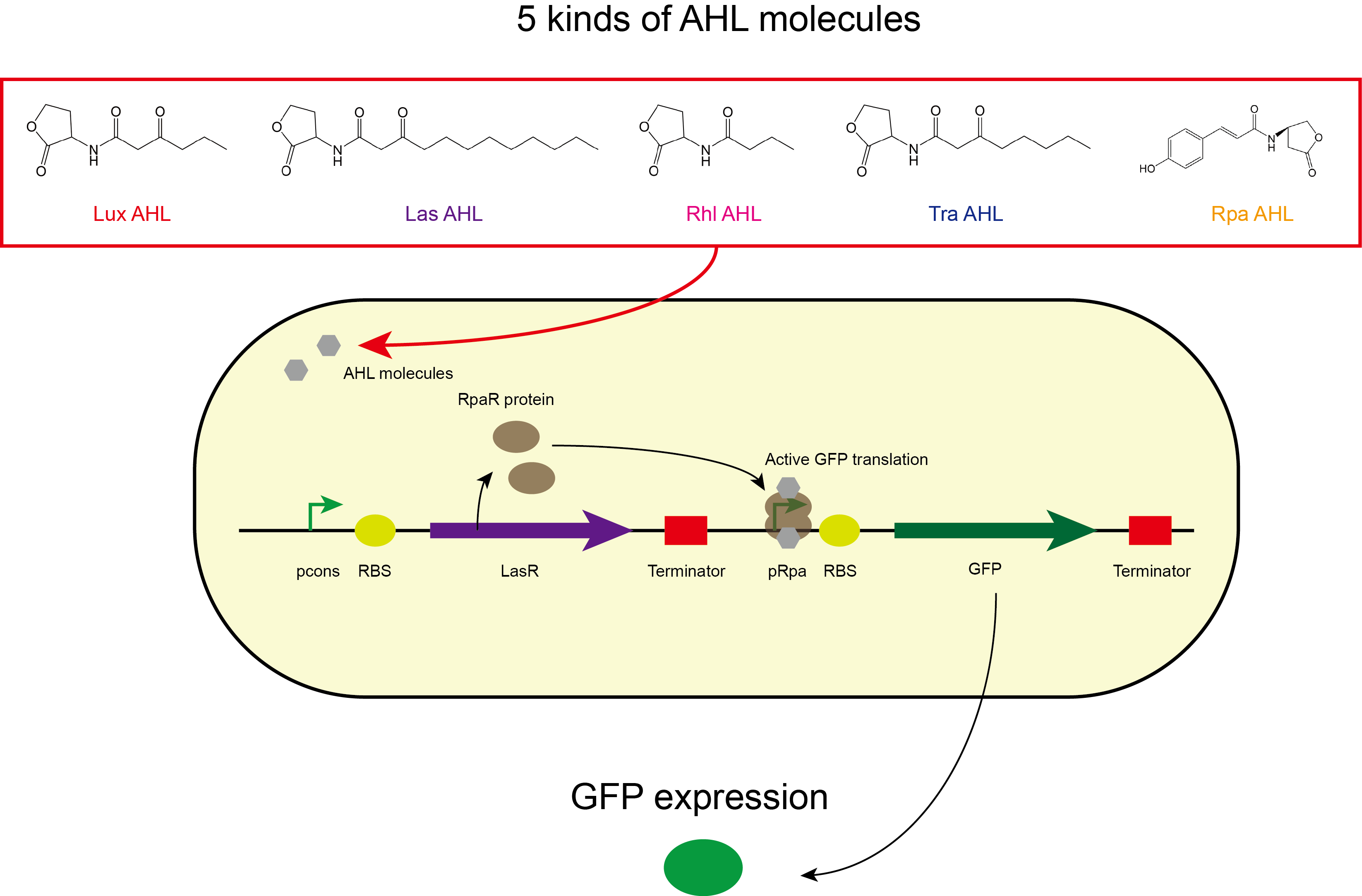
We not only found the activation range of LasR when induced with3OC12-HSL, but also tested the other 4 kinds of AHLs. We placed the coding sequence downstream from the pLas promoter, and recorded the fluorescence for different concentrations of AHLs. We found that there was no significant activation of LasR by AHLs of Lux、Rhl, however, Tra has a remarkable crosstalk.
- (the circuit design (.dna file) can be download in the part’s ‘design page’; the characterization of them can be found in the part’s ‘experiment page’.)
I. Characterisation of LasR by Imperial College London iGEM 2016
Group: Imperial College London 2016
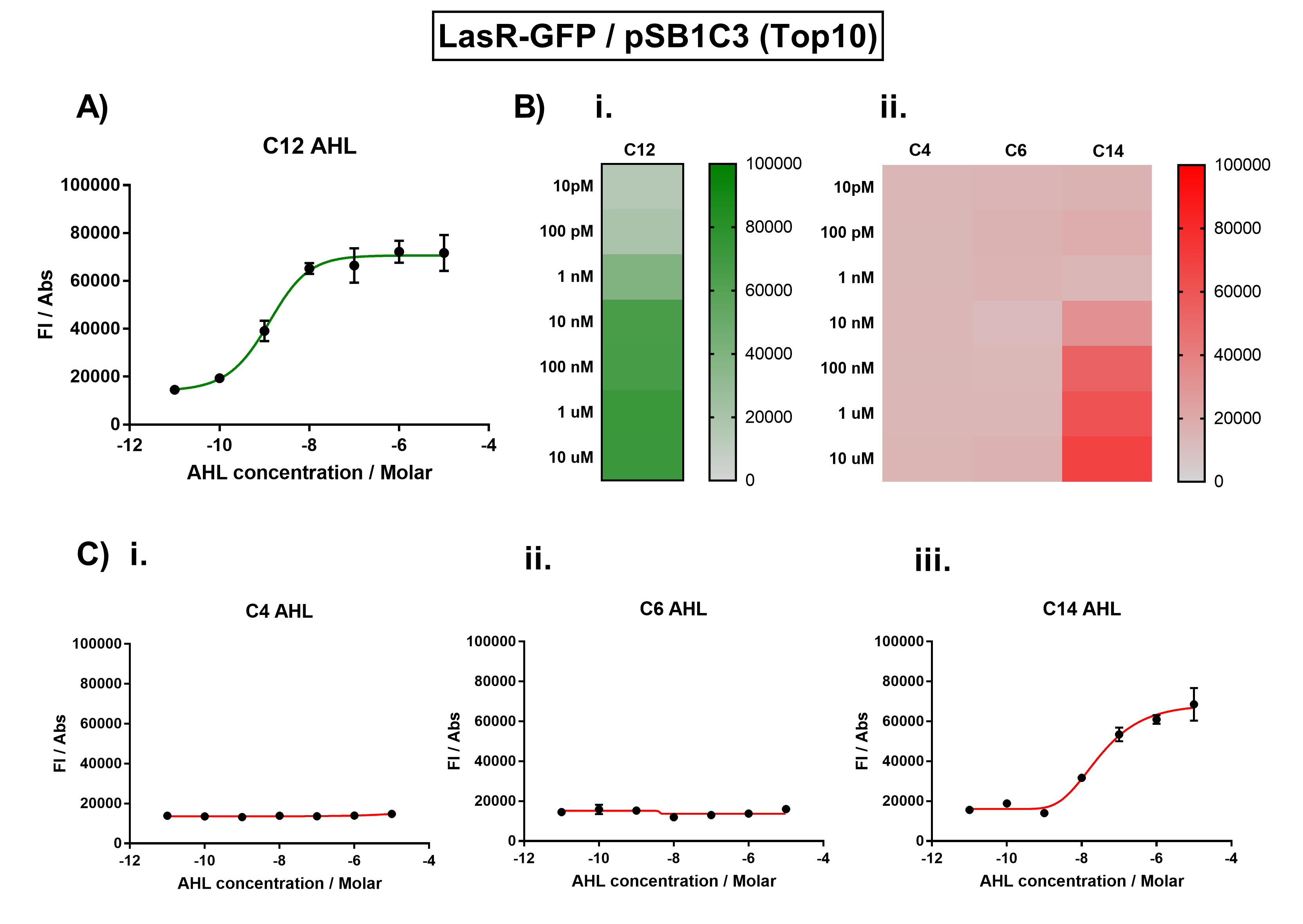
We improved this part by characterising the crosstalk between LasR and non-cognate AHLs. We found the activation range of LasR when induced with C12-AHL. For the crosstalk experiments, we tested C4-AHL, C14-AHL, and C6-AHL. We placed the coding sequence downstream from the pLas promoter, and recorded the fluorescence for different concentrations of AHLs.
We found that there was no significant activation of LasR by the C4-AHL.
Figure 1. Characterisation of the Las response device (BBa_K1893001). (A) Transfer function curve of normalised fluorescence against cognate inducer C12-AHL (3O-C12 AHL) concentrations. (B) Heat map of normalised fluorescence of RhlR-GFP system over a range of AHL concentrations: (i) Binding of RhlR-GFP to its cognate AHL (C4 AHL). (ii) Binding of RhlR-GFP to 3 non-cognate AHLs (3O-C6 AHL, 3O-C12 AHL, 3OH-C14 AHL). (C) Transfer function curves of normalised fluorescence against non-cognate inducer AHL (3O-C12 AHL) concentrations to investigate inducer AHL crosstalk: (i) C4-AHL of the Rhl system (ii) C6-AHL (3O-C6 AHL) of the Lux system (iii) C14-AHL (3O-C14 AHL) of the Cin system. Experiments were performed in E. coli Top10 cell strain cultured at 37°C. Normalised fluorescence was calculated by dividing fluorescent signal by cell density (OD600). Fluorescence measurements were recorded at 180 minutes. Reported values represent the mean normalised fluorescence value from 3 technical repeats and error bars represent standard deviation of these.
HKGTC 2023
Group: HKGTC 2023
In our project, transcription factor LasR was expressed in the sensing module, which can bind to the synthetic AHL molecules to form LasR-AHL complex. The complex can then bind to inducible promoter, pLasRL to activate the EGFP production. BBa_C0179 was used as the LasR transcription factor in the sensing module of our composite part. Our results showed that in the presence of the 1x10-7M AHL molecules, C10HSL and 3OC12-HSL, the engineered biosensor cells expressing LasR in the sensing module produced strong green fluorescence signals while in the presence of C4HSL and in the control set-up, there was no green fluorescence signals observed. This showed that the LasR sensing module was able to detect C10HSL and 3OC12-HSL.

Fig. 1 Fluorescence microscope images of PSBIC3-LasR-pLasRL-EGFP (BBa_K4946004) incubated with individual synthetic AHL molecules (C4HSL, C10HSL and 3OC12-HSL) at a concentration of 1x10-7M in 3 hours.

Figure 2. EGFP production of PSBIC3-LasR-pLasRL-EGFP (BBa_K4946004) toward three individual synthetic AHL molecules at 1x10-7M C4HSL, C10HSL and 3OC12-HSL for 4h. Distilled water was used as a negative control.
The results demonstrated that EGFP production rate of the biosensor was the highest when it was incubated with 3OC12-HSL followed by C10HSL. The biosensor did not show response when it was incubated with C4HSL.
To conclude, our data showed that lasR activator from P. aeruginosa PAO1 (BBa_C0179) can detect AHL molecules, 3OC12-HSL and C10HSL.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 322
Illegal AgeI site found at 519 - 1000COMPATIBLE WITH RFC[1000]
//cds/transcriptionalregulator/activator
//function/cellsignalling
//function/regulation/transcriptional
| direction | Forward |
| function | Rep |
| ligands | PAI |
| protein | lasR |
| rbs | No |
| swisspro | P25084 |
| tag | None |

 1 Registry Star
1 Registry Star