Coding
Part:BBa_K2656018
Designed by: Adrián Requena Gutiérrez, Carolina Ropero Group: iGEM18_Valencia_UPV (2018-09-23)
amilCP Coding Sequence
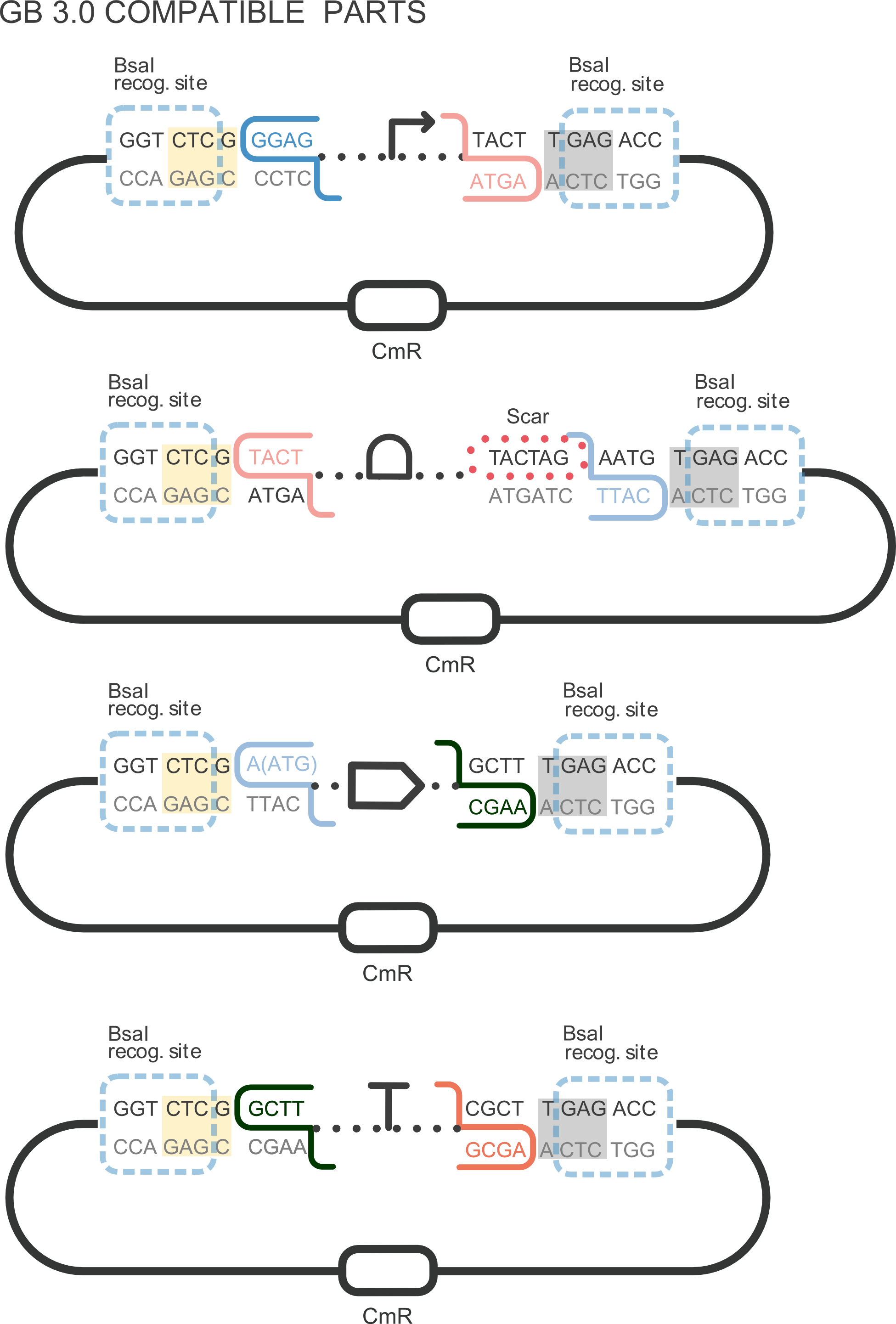
Part BBa_K2656018 is the amil Chromoprotein coding sequence BBa_K592009 compatible with both BioBricks and [http://2018.igem.org/Team:Valencia_UPV/Design Golden Braid 3.0] assemply methods. It can be combined with other compatible parts from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Valencia UPV IGEM 2018 Printeria Collection] to assemble transcriptional units with the Golden Gate assembly protocol .
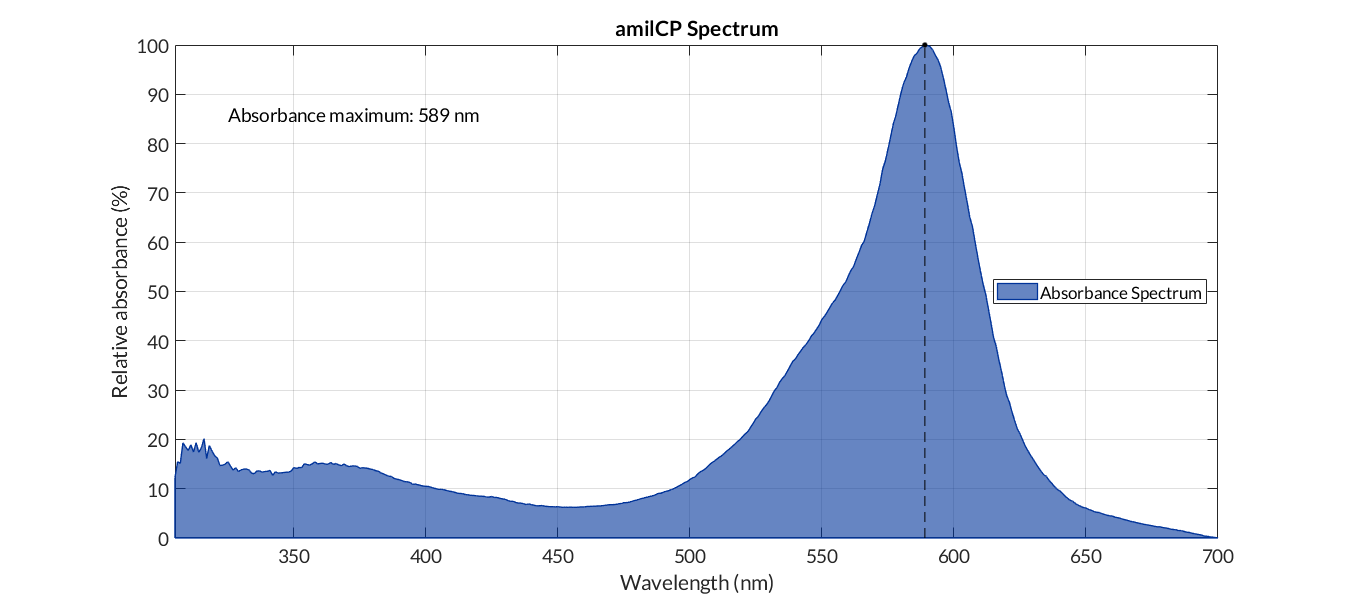
The characterization of this protein (and by extension of all the other part that codify for the amilCP) was performed with our transcriptional unit BBa_K2656113. This transcriptional unit was assembled in a Golden Braid alpha 1 plasmid including the following parts:
- BBa_K2656004: the J23106 promoter in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- BBa_K2656009: the B0030 ribosome biding site in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- BBa_K2656018: This part.
- BBa_K2656026: the B0015 transcriptional terminator in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
| Parameter | Value | ||
| Number of samples | 3 | ||
| Wavelength measurement range (nm) | [300-700] | ||
| Table 1. Parameters used to obtain the spectum | |||
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
[edit]
Categories
Parameters
//cds/reporter/chromoprotein
//function/reporter/pigment
//function/reporter/pigment
| None |