Part:BBa_K2656010
Weak Ribosome Binding Site B0032
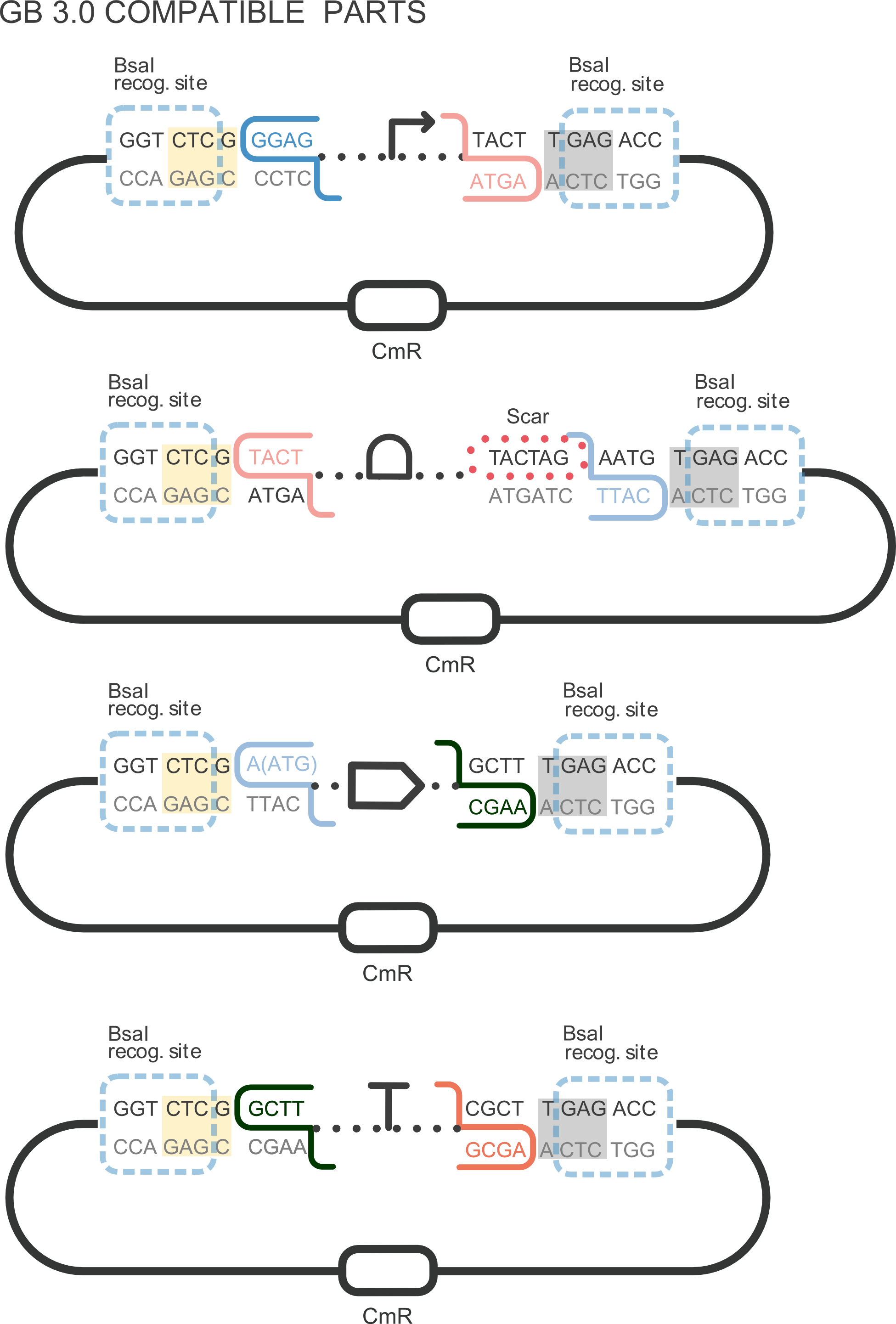
BBa_K2656010 is the BBa_B0032 ribosome binding site standardized into the Golden Braid assembly method. Thus, it is weak RBS compatible both with the BioBrick and Golden Gate grammar. It also includes the BioBrick equivalent scar in the 3' extreme, so the insertion of this supplementary bases bases ensure correct spacing for the CDS expression when assembled into a TU.
This RBS is part of our complete Printeria RBS Basic Part Collection
| Part | BioBrick | Description | Strength (Relative to BBa_K2656009) |
| BBa_K2656009 | BBa_B0030 | Strong RBS | 1 |
| BBa_K2656011 | BBa_B0034 | Medium RBS | 0.38 |
| BBa_K2656010 | BBa_B0032 | Weak RBS | 0.047 |
| BBa_K2656010 | BBa_J61100 | Very weak RBS | 0.042 |
| BBa_K2656012 | BBa_J61101 | Very weak RBS | 0.032 |
Using the Golden Gate assembly protocol , it can be combined with other GB adapted basic parts, such as the ones from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Valencia UPV IGEM 2018 Printeria Collection], to assemble transcriptional units in a single one-pot reaction.
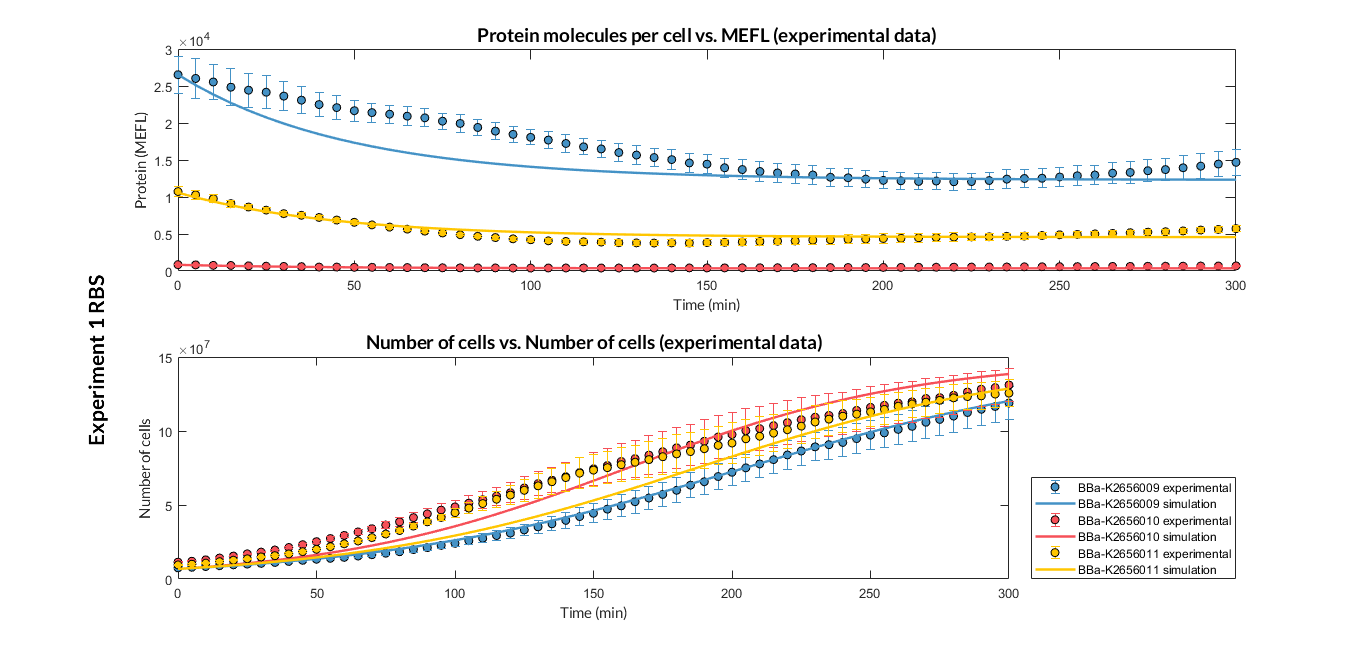
Characterization of this part was performed with the transcriptional unit BBa_K2656102, which was used in a comparative RBS expression experiment with composite parts BBa_K2656103 and BBa_K2656101.
They all were assembled in a Golden Braid alpha1 plasmid using the same promoter, coding sequence and terminator:
- Promoter BBa_K2656004: the J23106 promoter in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- CDS BBa_K2656013: the BBa_K2656009 sfGFP sequence in its Golden Braid standardized version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- Terminator BBa_K2656026: the B0015 transcriptional terminator in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model]and rationale choose its optimized values based on each RBS tested.
| Table 1. Optimized parameters for the BBa_K2656010 RBS. | |||
| Parameter | Value | ||
| Translation rate p | p = 0.01 min-1 | ||
| Dilution rate μ | μ = 0.01641 min-1 | ||
We have also calculated the relative force between the different RBS, taking BBa_K2656009 strong RBS as a reference. It has been defined as the quotient between the values of the protein in equilibrium of the results of the simulation of one RBS and another reference RBS. Likewise, a ratio between p parameters of the different RBS parts and p parameter of the reference RBS has been calculated.
| Table 2. BBa_K2656010 (GB B0030 RBS) relative strength and p ratio. | |||
| Parameter | Value | ||
| Relative strength | 0.045 | ||
| p parameter ratio (pRBS/pref) | 0.048 | ||
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//ribosome/prokaryote/ecoli
//chassis/prokaryote/ecoli
//direction/forward
//regulation/constitutive
| None |