Assembly standard 28
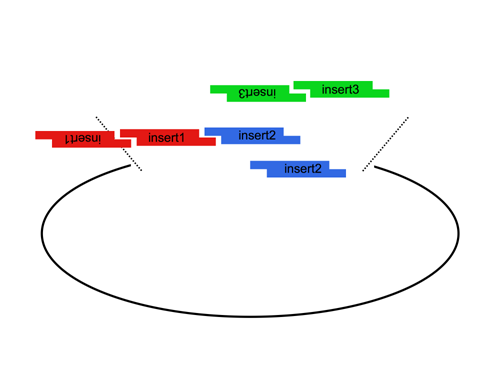
| The Lim standard is based on a multi-part/combinatorial cloning technique that is particularly well suited to shuffling protein domains. The key to this approach is the Type IIS restriction enzyme, AarI, a rare (7-cutter) that cuts 4bp offset from its binding site. Thus, AarI can generate four base overhangs of any sequence. | 
|
|
Since the user can specify the overhangs, this method can be used to "stitch-together" fragments without a scar, which is sometimes necessary to preserve protein function. More importantly, these overhangs can be non-palindromic thereby avoiding a key problem of trying to do multipart ligations using standard restriction enzymes: the self ligation of a part (which blocks proper assembly of parts). |
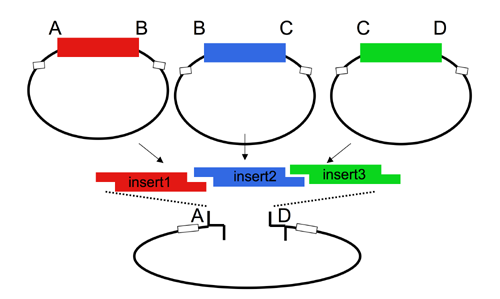
| The Lim standard enables high efficiency ligations of up to 4 parts simultaneously (vector plus 3 inserts). While parts can be made with any 4 base overhangs, we chose a standard set, termed A, B, C, and D to facilitate exchange of parts between researchers.
Note that most parts that adhere to the Lim standard are primarily intended for use when working with yeast. |
|
| Plasmid backbones (?) | Protein coding sequences (?) | Protein domains (?) |
Or get help on Assembly standard 28 parts.
Plasmid backbones
Plasmids are circular, double-stranded DNA molecules typically containing a few thousand base pairs that replicate within the cell independently of the chromosomal DNA. Plasmid DNA is easily purified from cells, manipulated using common lab techniques and incorporated into cells. Most BioBrick parts in the Registry are maintained and propagated on plasmids. Thus, construction of BioBrick parts, devices and systems usually requires working with plasmids.
Note: In the Registry, plasmids are made up of two distinct components:
- the BioBrick part, device or system that is located in the cloning site, between (and excluding) the Assembly standard 28 prefix and suffix.
- the plasmid backbone which propagates the BioBrick part. The plasmid backbone is defined as the sequence beginning with the Assembly standard 28 suffix, including the replication origin and antibiotic resistance marker, and ending with the Assembly standard 28 prefix. [Note that the plasmid backbone itself can be composed of BioBrick parts.]
Many BioBrick parts in the Registry are maintained on more than one plasmid backbone!
While any vector can be adapted to adhere to the Lim standard, most of the available vectors from the Registry that adhere to the Lim standard are derived from the yeast pRS__ series of vectors. The Registry has several types of acceptor vectors built in the pRS315 or 305 backbone. When necessary, markers can be exchanged by one-piece subclones of the completed cassettes into alternative pRS vectors, using the Kpn1/PspOMI and SacI sites.
There are no parts for this table
Protein coding sequences
Protein coding sequences are DNA sequences that are transcribed into mRNA and in which the corresponding mRNA molecules are translated into a polypeptide chain. Every three nucleotides, termed a codon, in a protein coding sequence encodes 1 amino acid in the polypeptide chain. In some cases, different chassis may either map a given codon to a different sequence or may use different codons more or less frequently. Therefore some protein coding sequences may be optimized for use in a particular chassis.
In the Registry, protein coding sequences begin with a start codon (usually ATG) and end with a stop codon (usually with a double stop codon TAA TAA). Protein coding sequences are often abbreviated with the acronym CDS.
Although protein coding sequences are often considered to be basic parts, in fact proteins coding sequences can themselves be composed of one or more regions, called protein domains. Thus, a protein coding sequence could either be entered as a basic part or as a composite part of two or more protein domains.
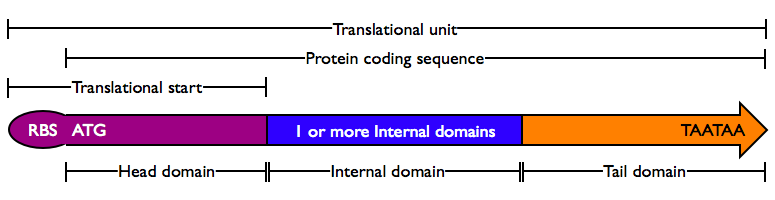
- The N-terminal domain of a protein coding sequence is special in a number of ways. First, it always contains a start codon, spaced at an appropriate distance from a ribosomal binding site. Second, many coding regions have special features at the N terminus, such as protein export tags and lipoprotein cleavage and attachment tags. These occur at the beginning of a coding region, and therefore are termed Head domains.
- A protein domain is a sequence of amino acids which fold relatively independently and which are evolutionarily shuffled as a unit among different protein coding regions. The DNA sequence of such domains must maintain in-frame translation, and thus is a multiple of three bases. Since these protein domains are within a protein coding sequence, they are called Internal domains. Certain Internal domains have particular functions in protein cleavage or splicing and are termed Special Internal domains.
- Similarly, the C-terminal domain of a protein is special, containing at least a stop codon. Other special features, such as degradation tags, are also required to be at the extreme C-terminus. Again, these domains cannot function when internal to a coding region, and are termed Tail domains.
For more details on protein domains including how to assemble protein domains into protein coding sequences, please see Protein domains.
Protein coding sequences should be as follows
GAATTC GCGGCCGC T TCTAG [ATG ... TAA TAA] T ACTAGT A GCGGCCG CTGCAG
There are no parts for this table
Protein coding sequences
Protein domains encode portions of proteins and can be assembled together to form translational units, a genetic part spanning from translational initiation (the RBS) to translational termination (the stop codon).
There are several different types of protein domains.
- Head Domain: The Head domain consists of the start codon followed immediately by zero or more triplets specifiying an N-terminal tag, such as a protein export tag or lipoprotein binding tag. Head domains should begin with an
ATGstart codon and include codons 2 and 3 of the protein at a minimum. Examples of head domains includeATGstart codonATGstart codon and codons 2-3ATGstart codon and signal sequenceATGstart codon and affinity tag
- Internal Domains: Protein domains consist of a series of codon triplets coding for an amino acid sequence without a start codon or stop codon. Multiple Internal Domains can be fused. Examples of internal domains include
- DNA binding domains
- Dimerization domains
- Kinase domains
- Special Internal Domains: Short Domains with specific function may be separately categorized, but obey the same composition rules as normal internal domains. Examples of special internal domains include
- Linkers
- Cleavage sites
- Inteins
- Tail Domain: The C-terminus of a coding region consists of zero or more triplet codons, followed by a pair of TAA stop codons. In the simplest case, the stop codons terminate the protein with an Stop. More complex Tails may include degradation tags appropriate to the organism (i.e., with different degradation rates). Examples of Tail domain include
TAATAAstop codons- A degradation tag followed by
TAATAAstop codons - An affinity tag followed by
TAATAAstop codon
Unfortunately, the original BioBrick assembly standard, Assembly standard 10, does not support in-frame assembly of protein domains. (Assembly standard 10 creates an 8 bp scar between adjacent parts.) Therefore, it is recommended that you use an alternate approach to assemble protein domains together to make a translational unit. There are several possible approaches to assembling protein domains including direct synthesis (preferred because it creates no scars) as well as various assembly standards. Regardless of which standard you choose, we suggest that the resulting protein coding sequence or translational unit comply with the original BioBrick assembly standard so that your parts can be assembled with most of the parts in the Registry.
Protein coding sequences should be as follows
GAATTC GCGGCCGC T TCTAG [ATG ... TAA TAA] T ACTAGT A GCGGCCG CTGCAG
Note: Although most RBSs are currently specified as separate parts in the Registry, we are now moving to a new design in which the RBS and Head domain are combined into a single part termed a Translational start. The new design has the advantage of encapsulating both ribosome binding and translational initiation within a single part. Our working hypothesis is that the new design will reduce the likelihood of unexpected functional composition problems between the RBS and coding sequence.
There are no parts for this table
References
<biblio>
- Lim Sergio G. Peisajovich, Andrew Horwitz, Oliver Hoeller, Benjamin Rhau & Wendell Lim. BBF RFC 28: A method for combinatorial multi-part assembly based on the Type IIs restriction enzyme AarI. BBF RFC 28. doi:1721.1/46721. [http://hdl.handle.net/1721.1/46721 DSpace].
- Sikorski pmid=2659436
</biblio>