Part:BBa_I746916
superfolder GFP coding sequence
This is the coding sequence of superfolder GFP (Pedelacq et al (2006): "Engineering and characterization of a superfolder green fluorescent protein", Nature Biotech 24 (1) January 2006).
More information about the properties of this GFP in relation to the currently used mut3GFP can be found at: http://openwetware.org/wiki/IGEM:Cambridge/2008/Improved_GFP.
It carries the following amino acid changes with respect to mut3 GFP (E0040), the currently most commonly used GFP in the registry:
S30R, Y39N, F64L, G65T, F99S, N105T, Y145F, M153T, V163A, I171V, A206V
Its in-vivo properties are considerably improved with respect to mut3 - it develops fluorescence about 3fold faster than mut3 GFP and reaches 4fold higher absolute fluorescence levels. Fluorescenct colonies can be identified with the naked eye even without UV or blue light illumination (that is to say the amount of blue light in normal daylight or lablight is sufficient). Additionally it is more stable in vitro and refolds faster after in vitro denaturation with respect to mut3 GFP.
Note: Superfolder GFP is available in constructs driven by the pBAD and T7 promoters: part numbers I746908 and I746909 respectively. Additionally 6-his tagged versions for protein purification exist: I746914 (pBAD driven) and I746915 (T7 driven).
Contribution: QHFZ-China 2019
Group: QHFZ-China iGEM 2019
Author: Cheng Li
Design:
This year we use BBa_K3007036 to express sfGFP. The part is equal to BBa_I746916. However, BBa_I746916 has two tandem termination codon (TGA) at the 3' end of the part. We retained only one termination codon (TGA) in BBa_K3007036.
Documentation:
We used charactered sfGFP part in the part BBa_K3007010 (https://parts.igem.org/Part:BBa_K3007010). Fig.4 and Fig. 5 showed the result related to sfGFP, Fig. 1-3 showed the process that we constructed the part.
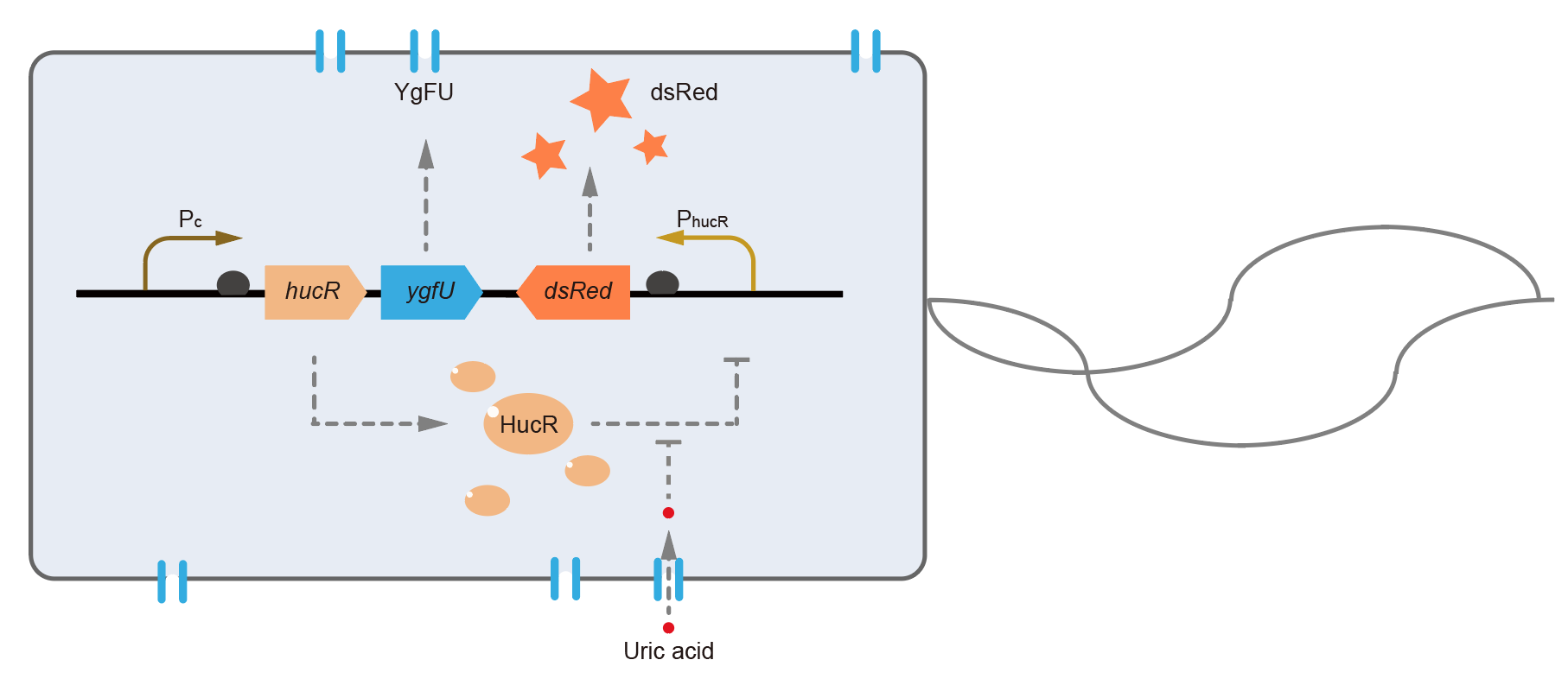
This year, QHFZ-China designed a UA monitor system in Escherichia coli (E. coli). The original version is shown in Fig. 1. Pc is a constitutive promoter, Pcp6 promoter, and it promotes the expression of HucR and YgfU. When uric acid is absent, HucR can bind to PhucR, which suppresses dsRed expression. If uric acid presents in high concentration, HucR will release from PhucR and the expression of dsRed will recover from the inhibition [2].
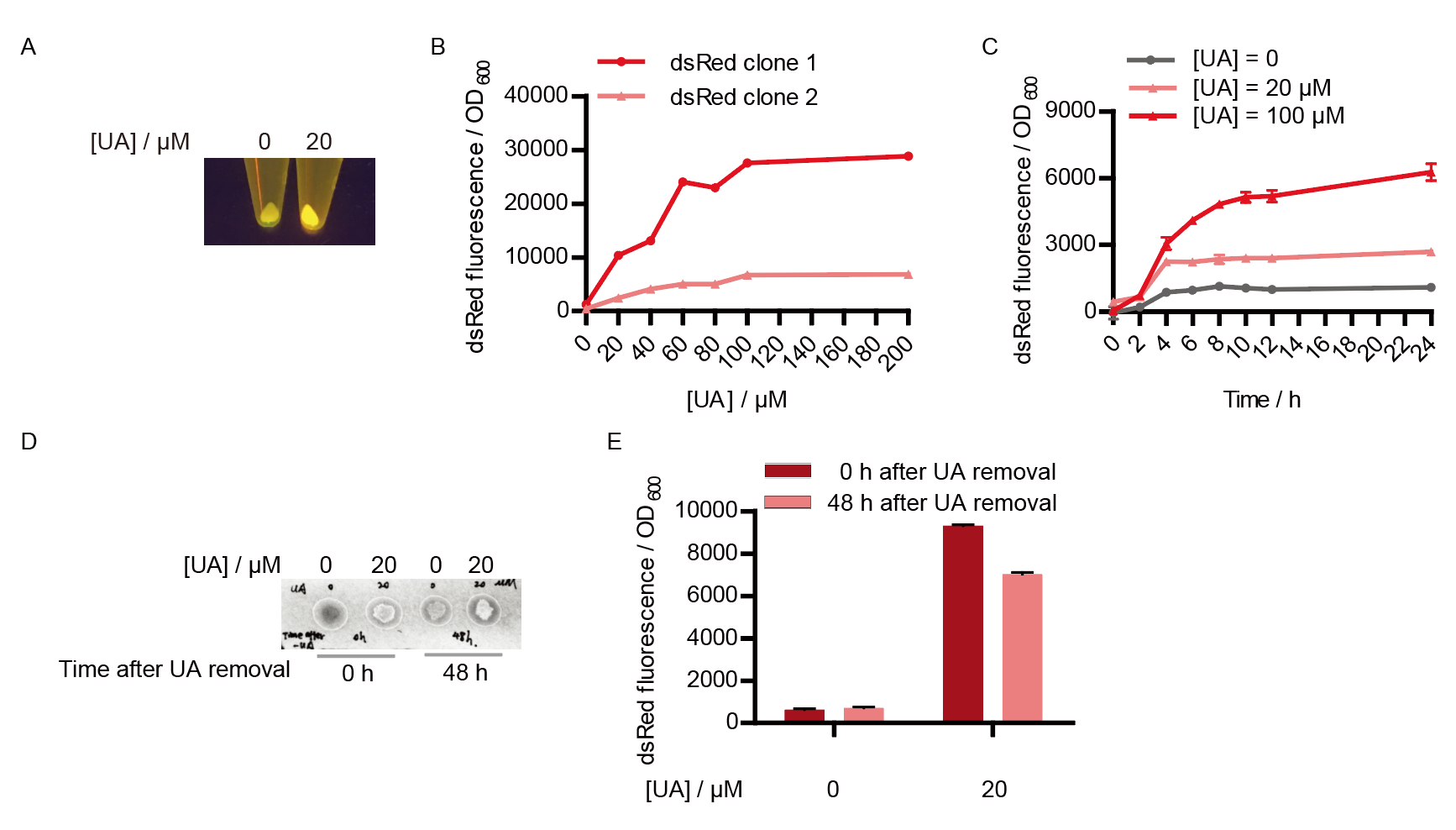
Two clones with the UA detection system were tested. The original gene circuit was able to response to UA in a range of 0 to 200 μM (Fig. 2A). The clone 1 showed much better dynamics than the other (Fig. 2B). Time course experiments showed that the fluorescence intensity became quite strong at 4 to 6 hours after UA induction, and became stable at 10 to 12 hours (Fig. 2C). Even if we removed UA by replacing fresh LB medium, after 48 hours shaking, the fluorescence would be still notable (Fig. 2D) and there was not significant difference of dsRed fluorescence / OD600 between before and after UA removing (Fig. 2E). All the data meant our design could detect high UA concentration quickly and stably.
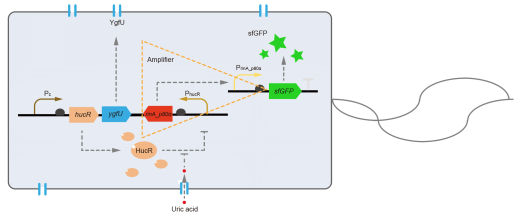
However, through our human practices, we found the sensitivity and responding time of the original design are not good enough. In the next generation design, we introduced RinA_p80α - PrinA_p80a system to enhance the sensitivity. Meanwhile, we changed dsRed to sfGFP, whose maturation time is much shorter, to shorten the waiting time. The new version of the uric acid detector was shown in Fig. 3. If UA presented, RinA_p80α would express and active transcription of sfGFP which was under control of PrinA_p80α. We called this as Version 2.
We tested the sfGFP production of Version 2 under different concentration of extracellular UA. The curve in Fig. 4A showed the fluorescence was saturated under only 15 μM UA induction, while the old version needed about 100 μM UA to get saturated (Fig. 2B). To test if sfGFP could shorten the reaction time, we used the same construct only except reporter genes, called PRinA_p80α – sfGFP and PRinA_p80α – dsRed, respectively. After adding 20 μM UA into the reaction system, the curve of PRinA_p80α – sfGFP climbed much faster than PRinA_p80α – dsRed, which suggested our new design had a great induction performance, and fitted our predictions very well (Fig. 4B).

At last, we took a photo to show the green fluorescence released by E. coli expressing sfGFP.
References:
[1] Wan, X., Volpetti, F., Petrova, E., French, C., Maerkl, S. J., & Wang, B. (2019). Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nature chemical biology, 15(5), 540.
[2] Liang C., Xiong D., Zhang Y., Mu S. and Tang S. (2015). Development of a novel uric-acid-responsive regulatory system in Escherichia coli. Appl. Microbiol. Biotechnol. 99, 2267–2275.
Contribution: WHU-China 2019
Group: WHU-China
Authors: Jiongyi He
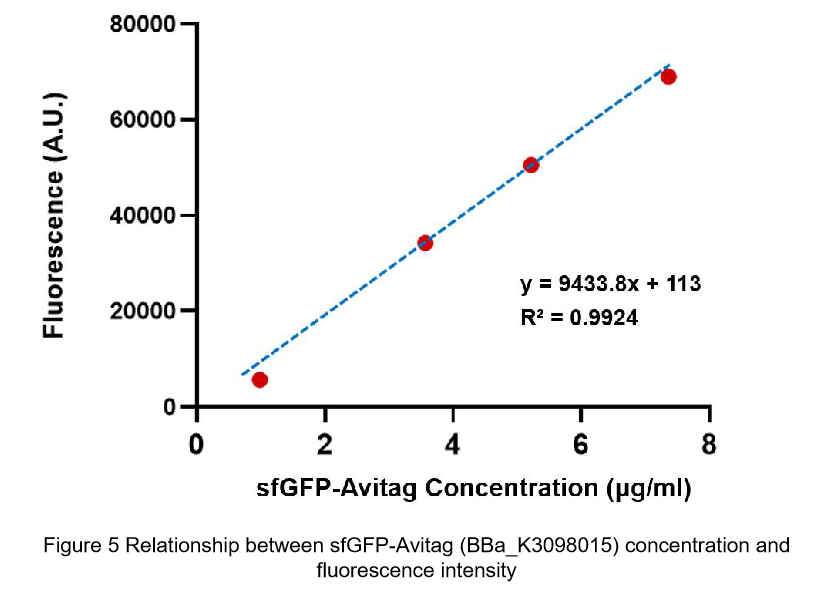
Summary: We confirmed that sfGFP (BBa_I746916) is available in constructs driven by the pR (BBa_R0051) and determined the relationship between sfGFP protein concentration and fluorescence intensity. Besides, we made a comparison fluorescence intensity of between BBa_K3098015 and BBa_I746916.
Documentation:

We constructed a recombined plasmid composed of pR (BBa_R0051)+RBS (BBa_B0030)+sfGFP (BBa_I746916)+6xHis-tag to express the sfGFP (BBa_I746916). By the way, there was not repressor CI of pR in the expression system.
We also constructed a recombined pet28a plasmid composed of T7 promoter and sfGFP <a href="https://parts.igem.org/Part:BBa_K3098015 ">BBa_K3098015</a> to express the sfGFP with avi-tag.
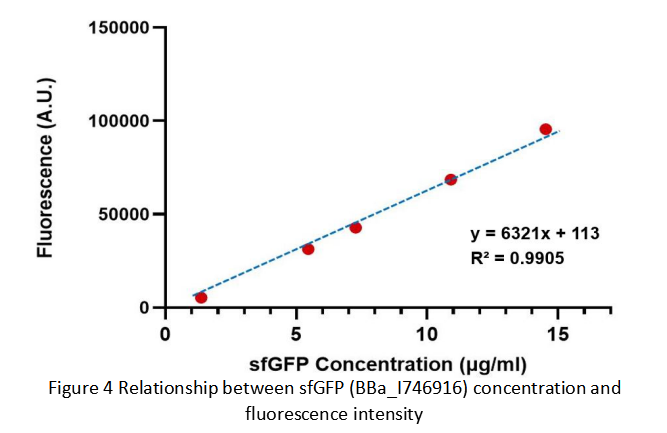
After purification by Ni resin, we made it a series of gradient dilutions of BBa_K3098015 and BBa_I746916 respectively. Then we measured the sfGFP protein concentration by Bradford method and the fluorescence intensity under 475nm as emission wavelength and 545nm as excitation wavelength.
Due to the fluorescence intensity of blank group without any sfGFP is 113 A.U., the ordinate at the origin should be 113. Then we fit data into a straight line: y=kx+b. It is easy to understand that the higher the value of k is, the stronger the sfGFP fluorescence is.
Therefore, we draw following conclusions:
(1) sfGFP (BBa_I746916) is available in constructs driven by the pR(BBa_R0051);
(2)According to the k of relationship between protein concentration and fluorescence intensity through our measurements, the fluorescence of sfGFP-Avitag (BBa_K3098015) is stronger than sfGFP (BBa_I746916).
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez
Summary: We have adapted the part to be able to assemble transcriptional units with the Golden Gate method and we have done the characterization of this protein.
Documentation:
We adapted the CDS BBa_I746916 to be used to assemble composite parts using the Golden Gate method creating BBa_K2656013. Then we made the characterization of the part:
The characterization of this protein (and by extension of all the other part that codify for the sfGFP) was performed with our transcriptional unit BBa_K2656101.
This transcriptional unit was assembled in a [http://2018.igem.org/Team:Valencia_UPV/Design Golden Braid alpha 1 plasmid] including the following parts:
- BBa_K2656004: the J23106 promoter in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- BBa_K2656009: the B0030 ribosome biding site in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- BBa_K2656013: This part.
- BBa_K2656026: the B0015 transcriptional terminator in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
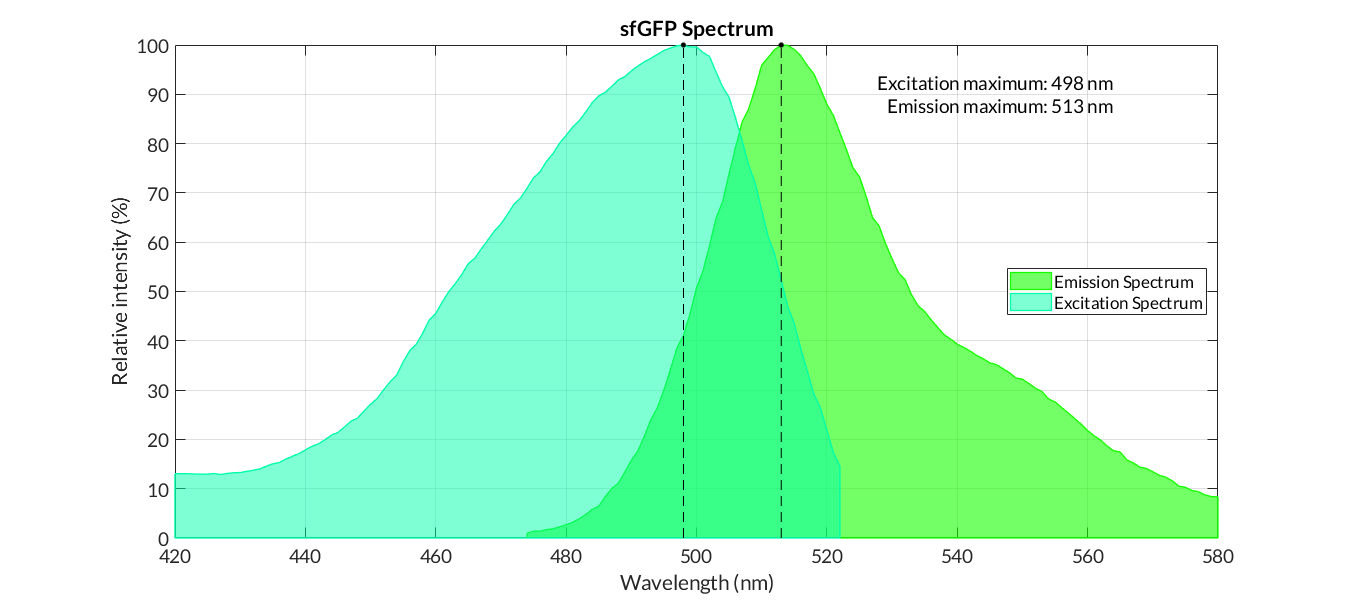
In order to carry out a correct characterization of the protein and to be able to use it to make measurements of the different transcriptional units that we have assembled with it, we have obtained the emission and excitation spectra in the conditions of our equipment. By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#spectra protocol] with the parameters of Table 1, Figure 1 has been obtained.
| Table 1. Parameters used to obtain the spectra | |||
| Parameter | Value | ||
| Number of samples | 6 | ||
| Excitation Wavelength measurement range (nm) | [420-525] | ||
| Emission wavelenght (nm) | 545 | ||
| Emission Wavelength measurement range (nm) | [470-580] | ||
| Excitation wavelenght (nm) | 450 | ||
| Gain (G) | 50 | ||
Improvement
Group: iGEM2018_Jilin_China
Author:Zihao Wang
Summary: This year our team registered the superfolder GFP designed by Overkamp W et al with a BBa_K2541400 (sfGFP_optimism, https://parts.igem.org/Part:BBa_K2541400). Compared with superfolder GFP(BBa_I746916), sfGFP_optimism (BBa_K2541400) is BbsI restriction site free, so it can be used in GoldenGate assembly to achieve efficient and rapid assembly of gene fragments.
Document:
1. Usage and Biology
Green fluorescent protein (GFP) exhibits intrinsic fluorescence and is commonly used as a reporter gene in intact cells and organisms [1]. Many mutants of the protein with either modified spectral properties, increased fluorescence intensity, or improved folding properties have been reported [2].
GFP often misfolds when expressed as fusions with other proteins, while a robustly folded version of GFP, called superfolder GFP (sfGFP), was developed and described by Pédelacq et al at 2006[3] that folds well even when fused to poorly folded polypeptides. We decided to use sfGFP as our reporter protein due to its faster folded feature and higher fluorescence intensity. The superfolder GFP had been registered in iGEM BBa_I746916. There is another superfolder GFP designed by Overkamp W et al at 2013[4], which is a codon optimized sfGFP. It was be used in Escherichia coli by Segall-Shapiro T H et al at 2018[5].
This year our team registered the superfolder GFP designed by Overkamp W et al with a BBa_K2541400 (called sfGFP_optimism) and BBa_K2541401(called sfGFP(BbsI free)).
Compared with superfolder GFP (BBa_I746916), sfGFP_optimism (BBa_K2541400) and sfGFP(BbsI free)(BBa_K2541401) are BbsI restriction site free, and the BbsI restriction endonuclease is an economical and efficient enzyme used in Golden Gate assembly, so sfGFP_optimism and sfGFP(BbsI free) can be used in Golden Gate assembly to achieve efficient and rapid assembly of gene fragments.
Figure 1. Expression of three types of sfGFP(BBa_I746916, BBa_K2541401, BBa_K2541400), cultivated overnight.
2. Characterization
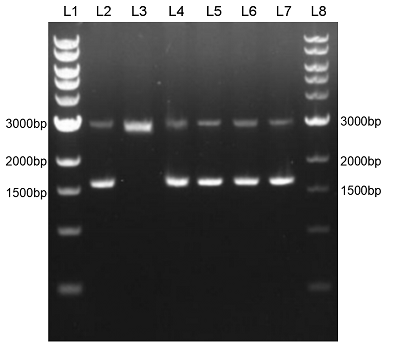
We constructed sfGFP_optimism (BBa_K2541400) sequence and superfolder GFP (BBa_I746916) on the pSB1C3 vector. The BbsI recognition site free was confirmed by nucleic acid electrophoresis (Figure 2). The sfGFP_optimism can not be digested by BbsI. And the length of these sequences are correct.
Figure 2. L1: 1kb DNA marker; L2: BBa_I746916; L3: BBa_I746916+BbsI; L4: BBa_K2541401; L5: BBa_K2541401+BbsI; L6: BBa_K2541400; L7: BBa_K2541400+BbsI; L8: 1kb DNA marker.
We got the emission and excitation spectra of two type sfGFP: sfGFP_optimism (BBa_K2541400), sfGFP(BbsI free)(BBa_K2541401) and superfolder GFP (BBa_I746916) (Figure 3).



Figure 3. Emission and Excitation Spectra of sfGFP_optimism(BBa_K2541400) and sfGFP(BBa_I746916)
For further characterization, we detected the expression intensity of these two types of sfGFP. According to the results (Figure 4), we found out that the fluorescence intensity of sfGFP_optimism (BBa_K2541400) is nearly 2.6 times higher than superfolder GFP (BBa_I746916).

Figure 5. The expression of three types of sfGFP in E.coli. The grey line represents fluorescent expression of sfGFP_optimism (BBa_K2541400), the orange line represents fluorescent expression of sfGFP(BbsI free) (BBa_K2541401), the blue line represents fluorescent expression of superfolder GFP (BBa_I746916) and the yellow line represents fluorescent expression of negative control (BBa_J364007).
3. Conclusion
We have made an improvement on the superfolder GFP (BBa_I746916). Our sfGFP_optimism (BBa_K2541400) is BbsI restriction site free, which can be used in Golden Gate assembly to achieve efficient and rapid assembly of gene fragments. And its fluorescence intensity is higher than superfolder GFP (BBa_I746916).
Contribution:UCAS-China 2019
Group:UCAS-China
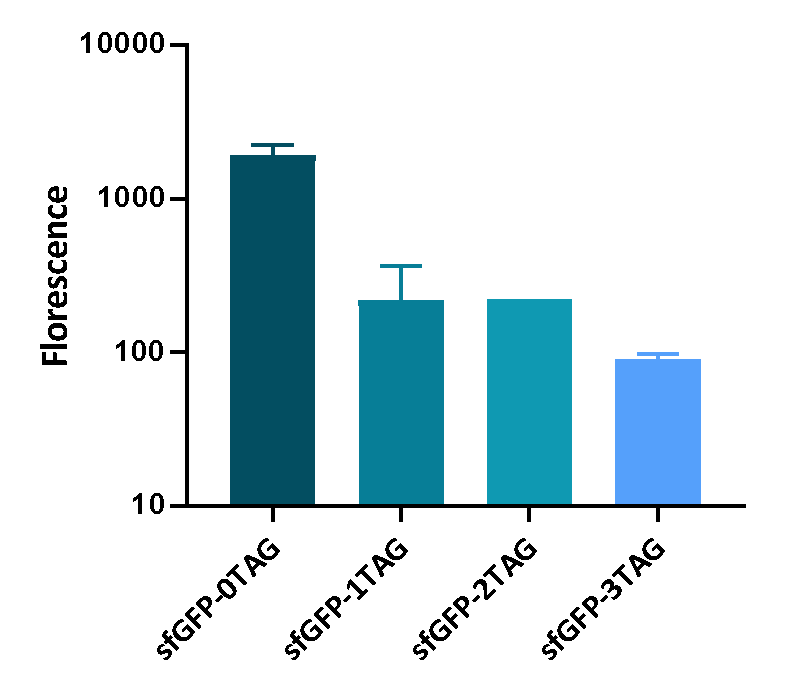
We inserted different number of TAG codon to sfGFP at different site to see the effect of TAG codon to the expression of sfGFP. Figure 1 shows that TAG codon can inhibit the transcription of sfGFP effectively.
Usage and Biology
Contribution: Linköping_Sweden 2019
Author: Andreas Holmqvist and Leo Juhlin
- BBa_B0034Ribosome binding site
- [http://https://parts.igem.org/Part:BBa_I719005 BBa_I719005]T7 promotor
Fluorescence in BL21 (DE3)
To verify the fluorescence of sfGFP (BBa_I746916), BL21 (DE3) containing CBD-sfGFP was grown in 1 liter LB-miller with 25 µg/ml chloramphenicol. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was used to induce the culture at a final concentration of 1 mM and the culture was incubated O.N. in 37 °C after the induction. Thereafter, the CBD-sfGFP expressing bacteria was placed on an UV-table emitting light 302 nm (Figure 5). The picture shows CBD-sfGFP´s strong fluorescence at 302 nm UV-light.
Compatibility in Vibrio natriegens
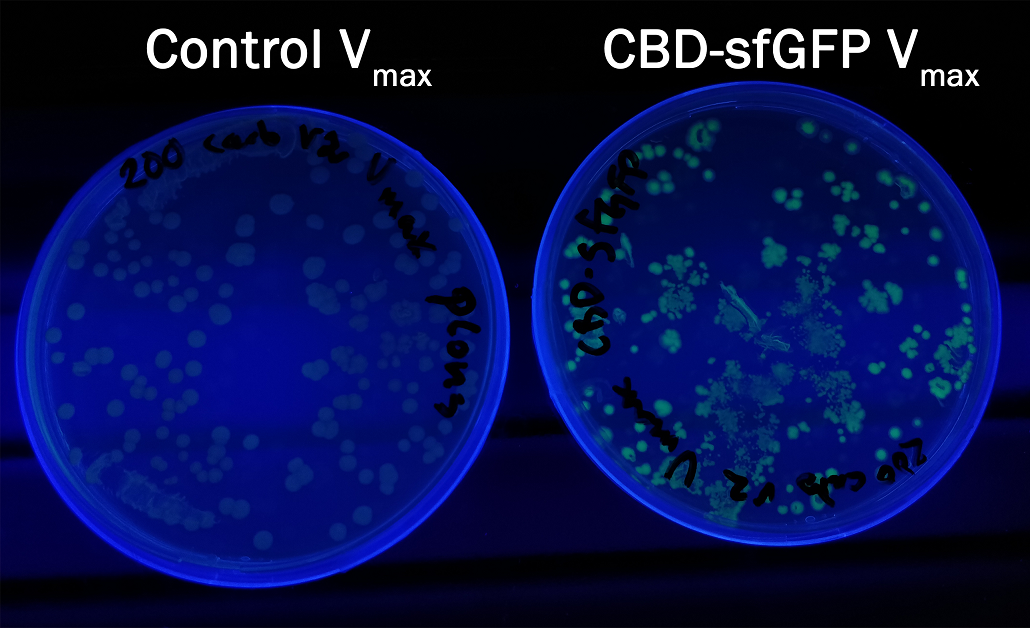
In order to see if sfGFP worked in Vibrio natriegens using the strain Vmax, CBD-sfGFP (BBa_K3182108) and CBD-pCons-Aspink (BBa_K3182100) was ligated into the pUC19 vector and heat shocked into Vmax.Thereafter, the bacteria was spread onto LB-miller V2 agar dishes with 200 µg/ml carbenicillin and incubated in 37 °C for 16 hours. Both plates was put on an UV-table and illuminated in 302 nm (Figure 6). The picture below shows that the CBD-sfGFP bacteria, in comparison to the control CBD-pCons-AsPink, displays a strong green fluorescent color which verified that pUC19-CBD-sfGFP could successfully be heat shocked and expressed in Vmax.
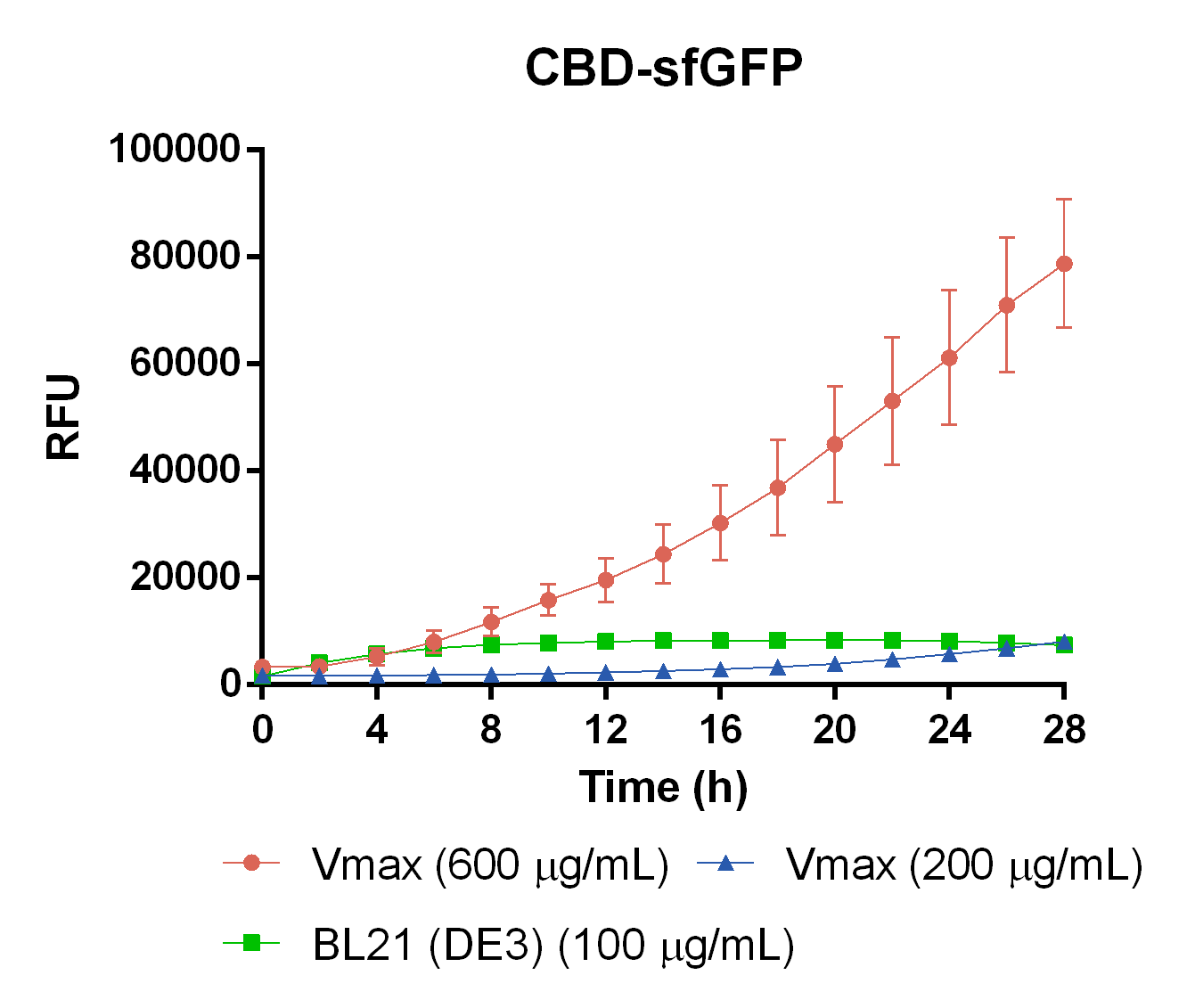
To measure the protein expression of T7-CBD-sfGFP in different bacteria and carbenicillin concentrations. BL21 (DE3) and Vibrio natriegens , using the strain Vmax, was grown in Falcon tubes to 0.5 OD600. Vmax was grown with two different carbenicillin concentrations, 200 and 600 µg/mL, while BL21 (DE3) had the same carbenicillin concentration of 100 µg/mL carbenicillin. The bacteria was induced with 1 mM IPTG and placed in a 96-well plate in 4 replicates with 200 µL per well. A spectrometry experiment was conducted and measured the fluorescence (excitation 470 nm,emission 550 nm) during 16 hours in 37 °C. The results seen below (Figure 7) shows that expression in Vmax with 600 µg/mL carbenicillin gave the highest protein yield. The most probable explanation for the increased protein yield for Vmax at 600 µg/mL carbenicillin is partially caused by the higher protein production of Vmax compared to BL21 (DE3). Another important factor was the use of an optimal concentration of carbenicillin (600 µg/mL) for Vmaxwhich retained the plasmid more efficiantly than Vmax at 200 µg/mL carbenicillin.
Contribution: Sydney_Australia 2019
Group: Sydney_Australia
Authors: Fahad Ali and Isobel Magrath
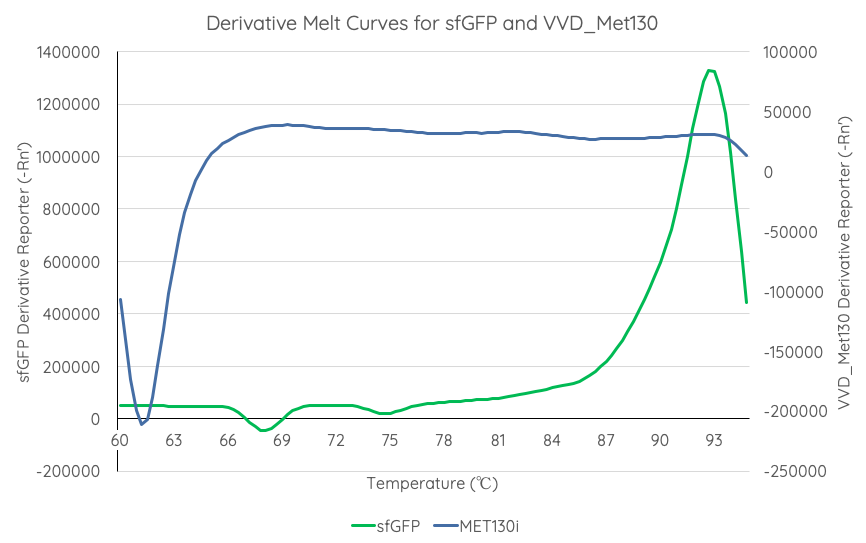
Summary: : We tested the thermal stability of sfGFP, by measuring fluorescence at nm over _____C using a qPCR machine.
Documentation:
</html>
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 13
| None |