Difference between revisions of "Part:BBa K2560010"
| Line 5: | Line 5: | ||
<html> | <html> | ||
<p align="justify">This is the Phytobrick version of the ribosome binding site <a href="https://parts.igem.org/Part:BBa_B0032">BBa_B0032</a> and was build as a part of the Marburg Collection. Instructions of how to use the Marburg Collection are provided at the bottom of the page. </p> | <p align="justify">This is the Phytobrick version of the ribosome binding site <a href="https://parts.igem.org/Part:BBa_B0032">BBa_B0032</a> and was build as a part of the Marburg Collection. Instructions of how to use the Marburg Collection are provided at the bottom of the page. </p> | ||
| + | </html> | ||
| + | |||
| + | ===Overview=== | ||
| + | |||
| + | <html> | ||
| + | <p align="justify"> | ||
| + | |||
| + | The ribosome binding site (RBS) is a short sequence upstream the transcription start. It can be identified easily by the typical Shine-Dalgarno sequence that pairs with the anti Shine-Dalgarno sequence at the 3’ end of the 16S rRNA | ||
| + | <a href=" https://www.ncbi.nlm.nih.gov/pubmed/7934914"><abbr title =" de Smit MH, van Duin J., Translational initiation at the coat-protein gene of phage MS2: native upstream RNA relieves inhibition by local secondary structure. (1993) 1079-88" >( Smit MH and Duin J van., 1993)</abbr></a>. | ||
| + | The effectiveness of this sequence is determined by its base-pairing potential to the ribosome and its spacing from the start codon | ||
| + | <a href=" http://www.sciencedirect.com/science/article/pii/S0022283605800245"><abbr title =" Maarten H. de Smit, Jan van Duin, Translational initiation on structured messengers: Another role for the shine-dalgarno interaction (1994) 173-184" >( Smit MH and Duin J van., 1994)</abbr></a>. The presence of the RBS is crucial for protein translation. | ||
| + | |||
| + | </p> | ||
</html> | </html> | ||
Revision as of 10:52, 17 October 2018
Phytobrick version of BBa_B0032
This is the Phytobrick version of the ribosome binding site BBa_B0032 and was build as a part of the Marburg Collection. Instructions of how to use the Marburg Collection are provided at the bottom of the page.
Overview
The ribosome binding site (RBS) is a short sequence upstream the transcription start. It can be identified easily by the typical Shine-Dalgarno sequence that pairs with the anti Shine-Dalgarno sequence at the 3’ end of the 16S rRNA ( Smit MH and Duin J van., 1993). The effectiveness of this sequence is determined by its base-pairing potential to the ribosome and its spacing from the start codon ( Smit MH and Duin J van., 1994). The presence of the RBS is crucial for protein translation.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Marburg Toolbox
We proudly present the Marburg Collection, a novel golden-gate-based toolbox containing various parts that are compatible with the PhytoBrick system and MoClo. Compared to other bacterial toolboxes, the Marburg Collection shines with superior flexibility. We overcame the rigid paradigm of plasmid construction - thinking in fixed backbone and insert categories - by achieving complete de novo assembly of plasmids.
36 connectors facilitate flexible cloning of multigene constructs and even allow for the inversion of individual transcription units. Additionally, our connectors function as insulators to avoid undesired crosstalk.
The Marburg Collection contains 123 parts in total, including:
inducible promoters, reporters, fluorescence and epitope tags, oris, resistance cassettes and genome engineering tools. To increase the value of the Marburg Collection, we additionally provide detailed experimental characterization for V. natriegens and a supportive software. We aspire availability of our toolbox for future iGEM teams to empower accelerated progression in their ambitious projects.
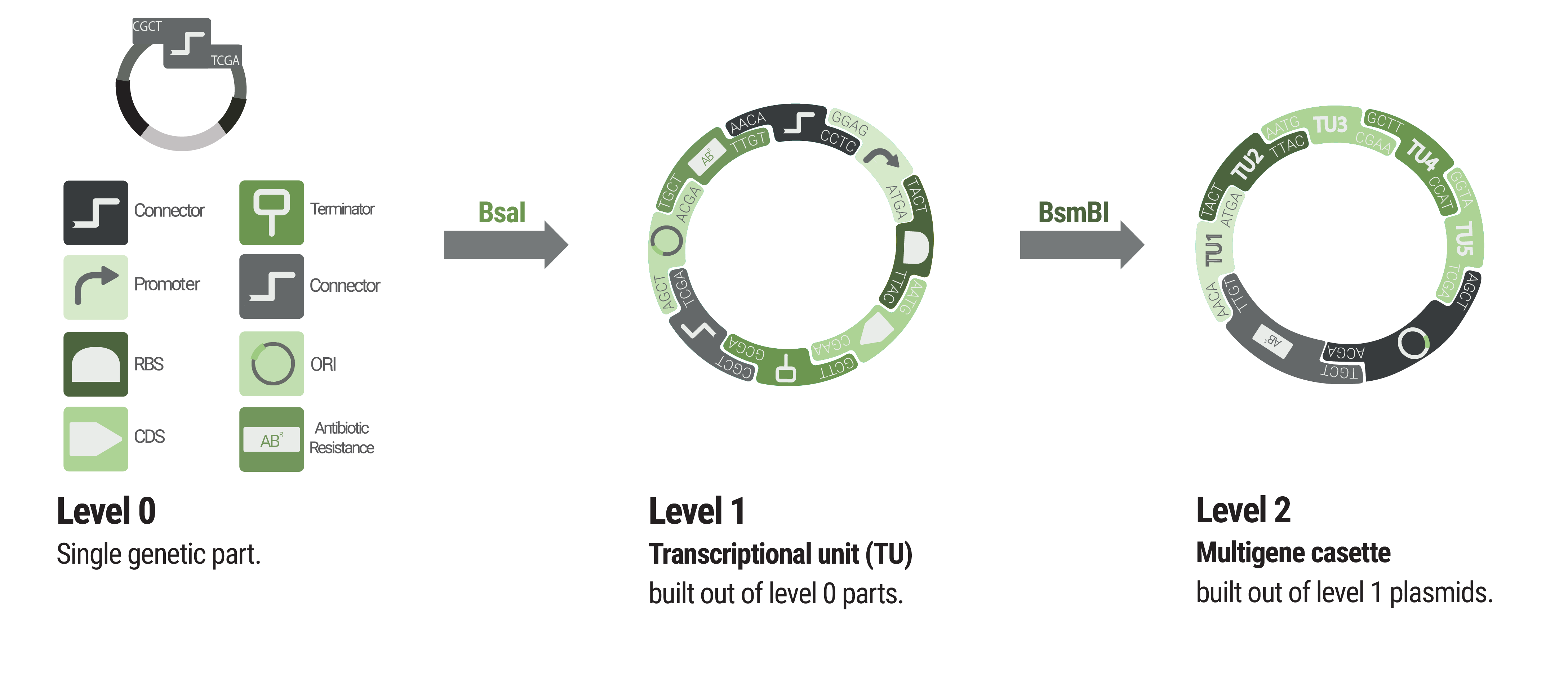
Basic building blocks like promoters or terminators are stored in level 0 plasmids. Parts from each category of our collection can be chosen to built level 1 plasmids harboring a single transcription unit. Up to five transcription units can be assembled into a level 2 plasmid.
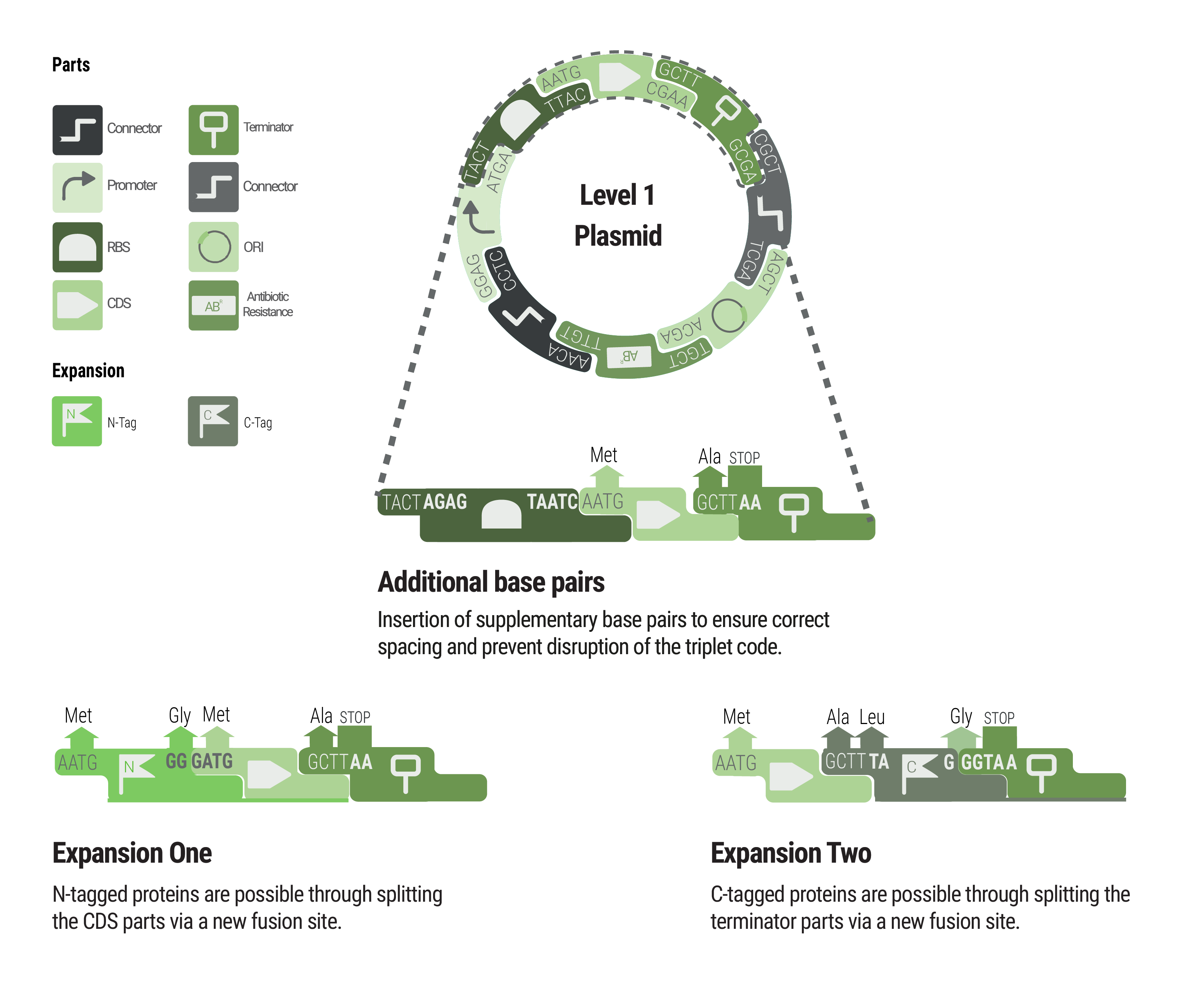
Between some parts, additional base pairs were integrated to ensure correct spacing and to maintain the triplet code. We expanded our toolbox by providing N- and C- terminal tags by creating novel fusions and splitting the CDS and terminator part, respectively.
Parts of the Marburg Toolbox

- K2560011 (5'Connector Dummy)
- K2560055
(1-6
Connector) - K2560065 (5'Con1)
- K2560066 (5'Con2)
- K2560067 (5'Con3)
- K2560068 (5'Con4)
- K2560069 (5'Con5)
- K2560075 (5'Con1
Short Res) - K2560076 (5'Con2
Short) - K2560077 (5'Con3
Short) - K2560078 (5'Con4
Short) - K2560079 (5'Con5
Short) - K2560095 (5'Con1 inv)
- K2560096 (5'Con2 inv)
- K2560097 (5'Con3 inv)
- K2560098 (5'Con4 inv)
- K2560099 (5'Con5 inv)
- K2560105 (5'Con5 inv
Ori) - K2560107 (5'Con1
Res)

- K2560007 (J23100)
- K2560009 (J23104)
- K2560014 (J23106)
- K2560015 (J23115)
- K2560017 (J23101)
- K2560018 (J23102)
- K2560019 (J23103)
- K2560020 (J23105)
- K2560021 (J23107)
- K2560022 (J23108)
- K2560023 (J23109)
- K2560024 (J23110)
- K2560025 (J23111)
- K2560026 (J23113)
- K2560027 (J23114)
- K2560028 (J23116)
- K2560029 (J23117)
- K2560030 (J23118)
- K2560031 (J23119)
- K2560123
(pTet) - K2560124 (pTrc)
- K2560131 (Promoter Dummy)

- K2560012 (3'Connector Dummy)
- K2560070 (3'Con1)
- K2560071 (3'Con2)
- K2560072 (3'Con3)
- K2560073 (3'Con4)
- K2560080 (3'Con5 Ori)
- K2560100 (3'Con1 inv
Short) - K2560101 (3'Con2 inv
Short) - K2560102 (3'Con3 inv
Short) - K2560103 (3'Con4 inv
Short) - K2560104 (3'Con5 inv
Short) - K2560106 (3'Con1 inv
Short Res) - K2560108 (3'Con1 inv)
- K2560109 (3'Con1 inv
Res) - K2560110 (3'Con2 inv)
- K2560111 (3'Con3 inv)
- K2560112 (3'Con4 inv)
- K2560113 (3'Con5 inv)

- K2560048 (Cam. Res. RFP)
- K2560056
(Kan. Res. (pSB3K3) RFP) - K2560057
(Kan. Res. (pSB3K3) GFP) - K2560058
(Tet. Res. (pSB3T5) RFP) - K2560059
(Tet. Res. (pSB3T5) GFP) - K2560125 (Carb. Res. RFP)
- K2560126 (Carb. Res. GFP)
- K2560127 (Carb. Res. into BBa_K2560002)
- K2560132 (Cam. Res. into BBa_K2560002)
- K2560133
(Kan. Res. into BBa_K2560002) - K2560134
(Tet. Res. into BBa_K2560002)
Tags and Entry Vectors







