Difference between revisions of "Help:Terminators"
(→Poly-A tails) |
m |
||
| (12 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | [[ | + | [[Category:Terminators]] |
| − | < | + | [[Terminators|< Back to Terminators]] |
| − | + | ||
| − | + | ||
| − | + | {{:Terminators/Overview}} | |
| − | + | *[[Help:Terminators/Mechanism|'''How''']] do terminators work? | |
| − | + | *[[Help:Terminators/Glossary|'''Glossary''']] of terms relating to terminators | |
| − | [[ | + | *[[Help:Terminators/Design|'''How to design a terminator''']] |
| − | + | *[[Help:Terminators/Construction|'''How to construct an terminator''']] | |
| − | + | *[[Help:Terminators/Measurement|'''How termination efficiencies are measured''']] | |
| − | + | *[[Help:Terminators/Further reading|'''Further reading''']] about terminator sequence and function | |
| − | + | *[[Terminators/Catalog|'''Browse''']] the terminator parts available from the registry | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 21:41, 20 July 2017
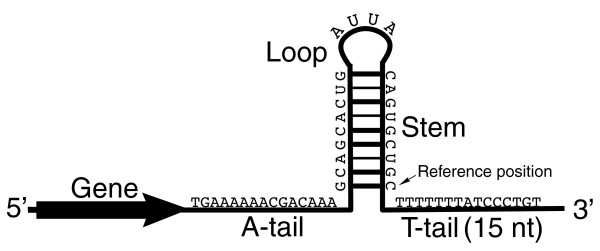
Terminators are genetic parts that usually occur at the end of a gene or operon and cause transcription to stop. In prokaryotes, terminators usually fall into two categories (1) rho-independent terminators and (2) rho-dependent terminators.
Rho-independent terminators are generally composed of palindromic sequence that forms a stem loop rich in G-C base pairs followed by several T bases. The conventional model of transcriptional termination is that the stem loop causes RNA polymerase to pause and transcription of the poly-A tail causes the RNA:DNA duplex to unwind and dissociate from RNA polymerase.
All the E. coli terminators in the Registry are rho-independent terminators. Rho-dependent terminators are not included, because rho-dependent terminators are not specified by sequence.
- How do terminators work?
- Glossary of terms relating to terminators
- How to design a terminator
- How to construct an terminator
- How termination efficiencies are measured
- Further reading about terminator sequence and function
- Browse the terminator parts available from the registry
