Help:Terminators/Mechanism
E. coli transcriptional termination
Terminators are genetic parts that usually occur at the end of a gene or operon and cause transcription to stop. In prokaryotes, terminators usually fall into two categories (1) rho-independent terminators and (2) rho-dependent terminators.
All the E. coli terminators in the Registry are rho-independent terminators. Rho-dependent terminators are not included, because rho-dependent terminators are not specified by sequence. 50% of the terminators following protein-coding regions and 70% of the terminators following non-coding RNAs are rho-independent terminators in E. coli Lesnik.
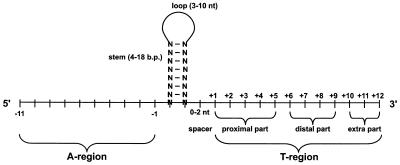
Rho-independent terminators are generally composed of palindromic sequence that forms a stem loop rich in G-C base pairs followed by several T bases. Transcription of a self-complementary region of the transcribed RNA causes a stem loop structure to form. Although the exact details are still unclear, the stem loop likely interacts with the E. coli RNA polymerase causing it to pause soon after the stem loop. Transcription of the poly-A tail results in an RNA:DNA duplex that is quite weak due to the low number of hydrogen bonds (2 hydrogen bonds for an U-A base pair vs 3 for a G-C base pair). The RNA:DNA duplex can thus unwind and dissociate from RNA polymerase. Transcription stops at U7 or U8 of the poly(T) tract.
Mutational analysis of rho-independent terminators shows that the stem loop sequence has a significant impact on the efficiency of termination Abe. The poly(T) tract also contributes to termination efficiency, but less so than the stem loop Abe.
Rho-independent terminators also impact gene expression by stabilizing the upstream mRNA transcript Abe; however, the poly(T) tract seems to play less of a role in mRNA stabilization Abe.
Eukaryotic transcriptional termination
Transcriptional termination in eukaryotes is less well understood than in bacteria Lodish. RNA polymerase I, II and III employ different termination mechanisms.
- RNA polymerase I terminates transcription of pre-rRNA genes with the help of a polymerase-specific termination factor that binds DNA. The termination factor binds downstream of the transcription unit.
- RNA polymerase II can terminate at multiple locations beyond the poly(A) addition site in most mammalian protein-coding genes. The locations can be 0.5-2 kb beyond the poly(A) addition site. Termination is coupled to the cleavage and polyadenylation of the transcript.
- RNA polymerase III terminates after transcribing a series of U residues. No upstream stem loop is necessary.
References
<biblio>
- Abe pmid=9150882
- Lesnik pmid=11522828
- Lodish isbn=0-7167-3706-x
</biblio>