Difference between revisions of "Part:BBa C0012"
Smelissali (Talk | contribs) |
RoyMustang (Talk | contribs) (→IMPROVEMENT REFERENCE: WHU-China 2022) |
||
| (50 intermediate revisions by 14 users not shown) | |||
| Line 4: | Line 4: | ||
[[Help:Protein coding|Coding region]] for the LacI protein with an LVA degradation tail and without an [[Help:Ribosome Binding Site|RBS]]. LacI binds to the pLac regulator <bbpart>BBa_R0010</bbpart> and PLlac01 hybrid regulator <bbpart>BBa_R0011</bbpart> and inhibits transcription. [http://openwetware.org/wiki/IPTG IPTG (Isopropylthiogalactoside)] binds to LacI and inhibits its operation, therefore promoting transcription. | [[Help:Protein coding|Coding region]] for the LacI protein with an LVA degradation tail and without an [[Help:Ribosome Binding Site|RBS]]. LacI binds to the pLac regulator <bbpart>BBa_R0010</bbpart> and PLlac01 hybrid regulator <bbpart>BBa_R0011</bbpart> and inhibits transcription. [http://openwetware.org/wiki/IPTG IPTG (Isopropylthiogalactoside)] binds to LacI and inhibits its operation, therefore promoting transcription. | ||
| − | A rapid degradation tail ([[Help:Tag|LVA]]) has been added to improve the High to Low performance of this part. | + | A rapid degradation tail ([[Help:Tag|LVA]]) has been added to improve the switch time for High to Low performance of this part. |
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | This particular LacI protein | + | This particular LacI protein was derived from ''E. coli'' and contributed by Michael Elowitz. (See Part Design for more information.) |
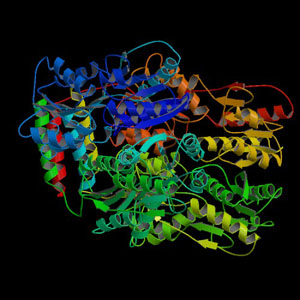
| − | [[Image:LacRep-black.jpg|frame|left|Image from the Protein | + | [[Image:LacRep-black.jpg|frame|left|Image from the Protein Data Bank (PDB - 1LBI)]] |
| + | |||
| + | **Allergen characterization of BBa_C0012: Not a potential allergen | ||
| + | |||
| + | The Baltimore Biocrew 2017 team discovered that proteins generated through biobrick parts can be evaluated for allergenicity. This information is important to the people using these parts in the lab, as well as when considering using the protein for mass production, or using in the environment. The allergenicity test permits a comparison between the sequences of the biobrick parts and the identified allergen proteins enlisted in a data base.The higher the similarity between the biobricks and the proteins, the more likely the biobrick is allergenic cross-reactive. In the full-length alignments by FASTA, 30% or more amount of similarity signifies that the biobrick has a Precaution Status meaning there is a potential risk with using the part. A 50% or more amount of identity signifies that the biobrick has a Possible Allergen Status. In the sliding window of 80 amino acid segments, greater than 35% signifies similarity to allergens. The percentage of similarity implies the potential of harm biobricks’ potential negative impact to exposed populations. For more information on how to assess your own biobrick part please see the “Allergenicity Testing Protocol” in the following page http://2017.igem.org/Team:Baltimore_Bio-Crew/Experiments | ||
| + | |||
| + | |||
| + | For the biobrick Part:BBa_C0012, there was a 0% of identity match and 0% similarity match to the top allergens in the allergen database. This means that the biobrick part is not of potential allergen status. In 80 amino acid alignments by FASTA window, no matches found that are greater than 35% for this biobrick. This also means that there is not of potential allergen status. | ||
| + | |||
| + | |||
| + | >Internal Priming Screening Characterization of BBa_C0012: Has 1 possible internal priming site between this BioBrick part and the VF2 primer. This BioBrick part also has 4 possible internal priming site between this part and the VR primer. | ||
| + | |||
| + | The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification. | ||
| + | |||
| + | For the BioBrick part BBa_C0012, the first location of the internal priming site is on the 207-214 base number of the BioBrick and on the 1-8 base number of the VF2 primer. The first location of the internal priming site is on the 628-622 base number of the BioBrick and on the 13-19 base number of the VR primer. The second location of the internal priming site is on the 947-941 base number of the BioBrick and on the 4-10 base number of the VR primer. The third location of the internal priming site is on the 778-784 base number of the BioBrick and on the 1-7 base number of the VR primer. The fourth location of the internal priming site is on the 1015-1021 base number of the BioBrick and on the 4-10 base number of the VR primer. | ||
| + | |||
| + | |||
| + | <br/> | ||
| + | <br/> | ||
| + | |||
| + | =Tsinghua 2018's characterization= | ||
| + | ==The Relation between LacI Concentration and IPTG Induction Efficiency== | ||
| + | ===I Background === | ||
| + | In order to investigate how lacI dosage affects IPTG induction, we used Anderson promotor J23100, J23110 and J23114 to design three constitutive lacI generators of different intensities. The three lacI generators were then ligated with Ptac driven reporter sfGFP to make three IPTG induction devices (BBa_K2558203, BBa_K2558204, BBa_K2558205). By measuring sfGFP fluorescence we tested how these devices react to IPTG. | ||
| + | <br/> | ||
| + | |||
| + | ===II Results=== | ||
| + | With high level of lacI expression (BBa_K2558203), sfGFP fluorescence had almost no response to IPTG induction. Weak lacI expression (BBa_K2558205) had the most significant IPTG induced sfGFP expression. With medium lacI expression level (BBa_K2558204), the induction efficiency lay in between. Therefore, the result proves that high level of lacI expression severely decreases IPTG induction efficiency [1]. Furthermore, IPTG concentration can affect the regulation part performance. The figure shows that without IPTG the sfGFP florescence intensity remained low. After IPTG addition, fluorescence signal immediately began to climb, forming a peak at five hours after induction, then sfGFP florescence intensity decreased and maintained at a lower level afterwards. IPTG concentration did not significantly affect the height of the peak or the expression level after the peak, but rather the peak width and expression stability of the system. Figures indicate that 5-10 mM IPTG had the most stable induction results. | ||
| + | |||
| + | <br/><li style="display: inline-block;"> [[File:T--Tsinghua-IPTGsfGFPresults-WithoutDescription.png|thumb|center|800px|'''Figure.1. The effect of varied lacI-LVA (BBa_C0011) promotor strength on IPTG induction of Tac promotor (BBa_K2558004).''' Relative fluorescent intensity is fluorescence per OD600 standardized with fluorescence per OD600 value of each test group at Time=0, IPTG=0. The promotors we used are from the Anderson collection: BBa_J23100 for strong lacI-LVA expression (pink), BBa_J23110 for medium expression (green), BBa_J23114 for weak expression (orange).]] | ||
| + | <!-- --> | ||
| + | |||
| + | ===III Protocol=== | ||
| + | # one Transform the plasmids into ''E. coli'' DH5α. | ||
| + | # two Pick a single colony by a sterile tip from each of the LB plates for all the experimental and control groups. Add the colony into 5ml M9 medium with ampicillin at 100 ng/µl. Incubate for 6-8 h at 37℃ in a shaker. | ||
| + | # three Measure OD600 of the culture medium with photometer. Dilute the culture medium until OD600 reaches 0.6. | ||
| + | # four Add 100 µl bacteria culture medium into a sterile 96-well plate. Add IPTG to final concentrations of 0, 1, 5, 10, 20 mM. Fresh M9 medium serves as blank control. Positive control is colony constantly expressing sfGFP and negative control is colony without sfGFP expression. Place the 96-well plate into an automatic microplate reader. Incubate at 16℃ overnight and record the fluorometric value at 510 nm and OD600 for each well every 30 minutes. | ||
| + | # five Each group should be repeated for at least 3 times. | ||
| + | |||
| + | ===IV Reference=== | ||
| + | [1] Szabolcs Semsey, Sandeep Krishna. "The effect of LacI autoregulation on the performance of the lactose utilization system in Escherichia coli" Nucleic Acids Res 2013 Jul; 41(13): 6381–6390 | ||
| + | |||
| + | <br/> | ||
| + | <br/> | ||
| + | =2019 Fudan-TSI's Improvement= | ||
| + | ==Overview== | ||
| + | This year team Fudan-TSI has upgraded LacI repressor coding gene to a new version (BBa_K3257045 https://parts.igem.org/Part:BBa_K3257045). | ||
| + | With lacIq promoter (BBa_K3257003 https://parts.igem.org/Part:BBa_K3257003) and rrnB T1 terminator (BBa_K3257020 https://parts.igem.org/Part:BBa_K3257020), improved LacI protein can be expressed and function properly in the Escherichia coli BL21(DE3). We used EGFP as a reporter controlled by our improved Lac operon and measured its green fluorescence over time. | ||
| + | |||
| + | For detailed design, please refer to https://parts.igem.org/Part:BBa_K3257045. | ||
| + | |||
| + | For a composite LacI repressor protein expression system, please refer to https://parts.igem.org/Part:BBa_K3257100. | ||
| + | |||
| + | According to our experiment, our Lac operon is improved in the following three main aspects. | ||
| + | |||
| + | ==Results== | ||
| + | ===Lower Response to Arabinose, Better Orthogonality=== | ||
| + | Cross talk between the response to IPTG and arabinose has been a defect of the wild type Lac operon. When 4mM arabinose added, a few lac inhibitors detach from lac operator which means that it is induced in a relatively low but unignorable level. According to the measurement of our experiment, our improved LacI can respond to IPTG with better orthogonality. The figure below (Figure 1) is the measurement of the fluorescence of EGFP controlled by wild-type and improved Lac operon. It shows that when 4mM arabinose is added, oLacI(wild-type LacI) is induced at a significantly high level while iLacI(improved LacI) is induced at a lower level. | ||
| + | [[File:ILacI Curves.png|center|500px|thumb|'''Figure 1. The expression level of EGFP controlled by different versions of LacI and inducers, or under different promoters.'''The origin point indicates the time when different inducers are added (1 mM IPTG and/or 4 mM Arabinose). The title of the graph shows which kind of inducer is added to the culture. The horizontal axis shows the duration of time, the vertical axis shows the quantified level of EGFP expression. The fluorescence level (excitation wavelength: 485 nm; detection wavelength: 528 nm) is quantified by the concentration of fluorescein, and normalized by the measured OD600 equivalent to the number of beads in the system. oLacI stands for the wildtype LacI, iLacI stands for our improved version of LacI. +LacO indicates that the promoter constitutes a LacO sequence. Control is the negative control plasmid which does not constitute an EGFP sequence. Error bar in the two graphs on the first row indicates the SEM of three replicates. The second row showed only the mean amount of three replicate.]] | ||
| + | ===Higher level of induction than the Wild-type Lac operon when Induced by IPTG=== | ||
| + | Next step we prove that our improved Lac operon can be better induced by IPTG, which counts a great deal for the usage of this operon. The figure below (Figure 2) is the measurement of the fluorescence of EGFP controlled by wild-type and improved Lac operon. It shows that when 1mM IPTG is added, EGFP controlled by both operons can be disinhibited and the improved LacI is induced at a relatively higher level. | ||
| + | [[File:ILacI+IPTG.png|center|500px|thumb|'''Figure 2. The induction level of EGFP under different repressors and promoters.''' The induction level is calculated by dividing the fluorescence level after 9 h of induction by 1 h afterward. The fluorescence level is quantified as in Fig. 1. oLacI stands for the wildtype LacI, iLacI stands for our improved version of LacI. +LacO indicates that the promoter constitutes a LacO sequence. t-test analysis shows that the induction level of iLacI is significantly higher than oLacI, *** indicates that p=0.0002.]] | ||
| + | |||
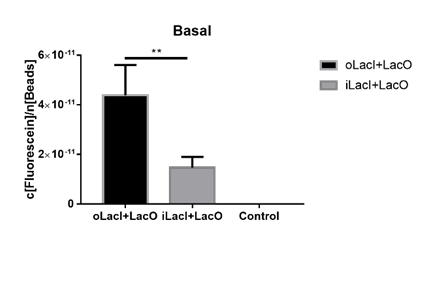
| + | ===Lower Uninduced Leakage=== | ||
| + | The uninduced leakage level is also an important parameter of an operon. Improved LacI lowers the leakage level compared to the wild-type one. The figure below (Figure 3) is the measurement of the fluorescence of EGFP controlled by wild-type and improved Lac operon. When no IPTG or arabinose is added, the fluorescence of EGFP controlled by improved Lac operon is under the detection range while the fluorescence of EGFP controlled by wild-type Lac operon remains a detectable signal indicating a considerably undesired leakage. | ||
| + | [[File:ILacI uninduced.png|center|500px|thumb|'''Figure 3. The basal fluorescence level of EGFP controlled by different repressors.''' The bar indicates the mean fluorescence level during the 10 h with no inducer in the culture. The fluorescence level is quantified as in Fig. 1. oLacI stands for the wildtype LacI, iLacI stands for our improved version of LacI. Control is below the detection level and not shown. Error bar indicates the SEM of fluorescence signal in the 10 h. Paired t-test analysis shows that iLacI has a significantly lower of fluorescence than oLacI, p=0.0057 (**).]] | ||
| + | |||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
| + | <partinfo>BBa_C0012 parameters</partinfo> | ||
| + | <h3><center>Burden Imposed by this Part:</center></h3> | ||
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 0.6 ± 2.1% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | ==References== | ||
| + | [1] Adam J. Meyer, Thomas H. Segall-Shapiro, Emerson Glassey, Jing Zhang & Christopher A. Voigt. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nature Chemical Biology volume 15, pages196–204 (2019) | ||
| + | |||
| + | <br/> | ||
| + | <br/> | ||
| + | =BHSF 2020's characterization= | ||
| + | This operator can not only be used in prokaryotic promoters, but also in eukaryotic promoters when inserted downstream of TATA-box. | ||
| + | |||
| + | After we decided to use toggle switch as the principle of our circuit, and use lac as a molecule in our circuit, we used these keywords to find the most suitable system for our yeast, and we found the article describing a hybrid promoter: | ||
| + | |||
| + | 1.If a lac operator site (lacO) is positioned appropriately downstream of the TATA‐box, gene expression from a nmt::lacO promoter (hybrid promoter) can be greatly repressed by lacI inhibitor. | ||
| + | |||
| + | 2.Insertion of the operator sequence into the nmt promoter 5 bp downstream of TATA-box causes irreversible repression of gene expression when lacI is present. | ||
| + | |||
| + | 3.When the operator in inserted directly downstream of TATA-box, the addition of IPTG to the medium could relieve the repression to some extent, but it is not fully reversible. | ||
| + | |||
| + | ==References== | ||
| + | Kjaerulff S, Nielsen O. An IPTG-inducible derivative of the fission yeast nmt promoter. Yeast. 2015 Jun;32(6):469-78. doi: 10.1002/yea.3073. Epub 2015 Mar 31. Erratum in: Yeast. 2016 Jan;33(1):33. PMID: 25801050. | ||
| + | |||
| + | |||
| + | |||
| + | <br/> | ||
| + | <br/> | ||
| + | |||
| + | =iBowu-China 2021's characterization= | ||
| + | |||
| + | ===I Design === | ||
| + | [[File:T--iBowu-China--2021lacI-1.png|thumb|center|800px|'''Figure.1. The plasmid design including lacI (BBa_C0011).''' ]] | ||
| + | |||
| + | <br/> | ||
| + | |||
| + | ===II Introduction=== | ||
| + | This year team iBowu-China used two parts LacI and LacO, with iPTG as induction methods, to control the expression of sfGFP protein and β-glucuronidase protein in E. coli BL21(DE3) | ||
| + | <br/> | ||
| + | |||
| + | ===III Protocol=== | ||
| + | # Transform the plasmids into ''E. coli'' BL21(DE3) | ||
| + | # Pick a single colony by a sterile tip from each of the LB plates for all the experimental and control groups. Add the colony into 4ml LB medium with kanamycin. Add 1mM iPTG to all experimental groups. Incubate at 37℃ in a shaker overnight. | ||
| + | # Add 100 µl bacteria culture medium into a sterile 96-well plate. Measure fluorescence at 488nm excitation and 502nm absorption, and then also OD600 for normalization, with a microplate reader. | ||
| + | <br/> | ||
| + | |||
| + | ===IV Results=== | ||
| + | '''sfGFP''' | ||
| + | [[File:T--iBowu-China--2021lacI-2.png|thumb|center|600px|'''Figure.2. The expression of sfGFP with lacI+sfGFP can be controlled by iPTG induction.''' ]] | ||
| + | Under the condition that there exists the part LacI, and there also exists LacO in the upstream gene of sfGFP, Iptg induction can control the switch on and off of the expression of green fluorescence protein sfGFP. | ||
| + | |||
| + | [[File:T--iBowu-China--2021lacI-3.png|thumb|center|600px|'''Figure.3. The expression of sfGFP with lacI+sfGFP can be switched on with different concentrations of Iptg induction.''' ]] | ||
| + | The plot and photo demonstrate that 0.5nM Iptg added into the medium is sufficient to induce the expression of sfGFP with the existence of lacI part. | ||
| + | |||
| + | '''Enzyme activity of β-glucuronidase''' | ||
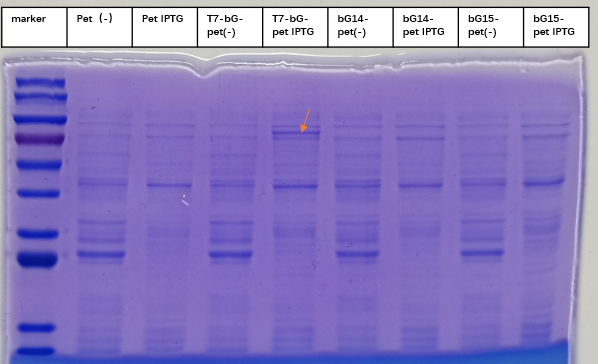
| + | [[File:T--iBowu-China--2021lacI-4.png|thumb|center|600px|'''Figure.4. The expression of β-glucuronidase | ||
| + | with lacI+sfGFP can be switched on with Iptg induction. The SDS-PAGE results clearly showed the expression of the enzynme at the indicated band. ''' ]] | ||
| + | |||
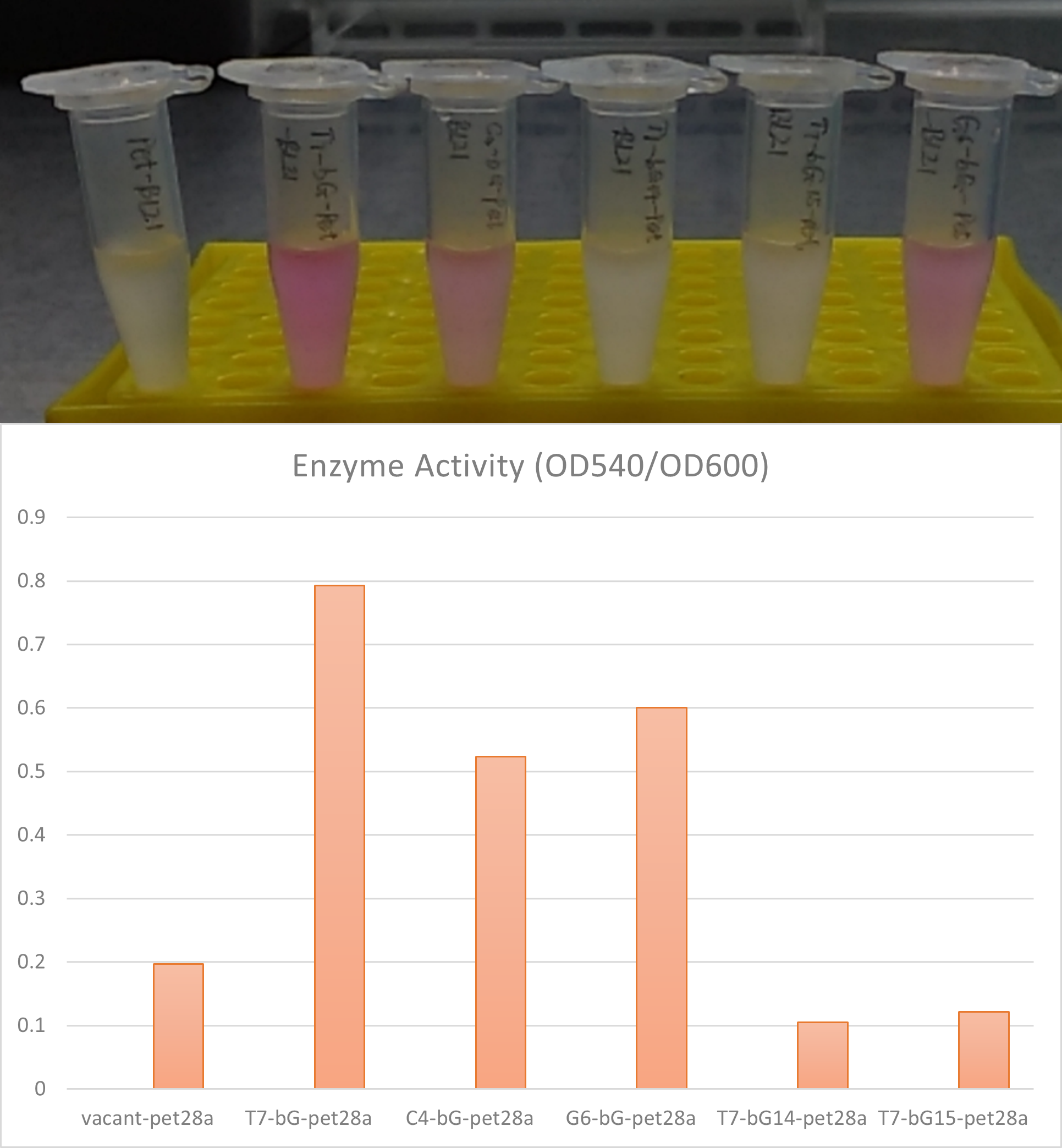
| + | [[File:T--iBowu-China--2021lacI-5.png|thumb|center|600px|'''Figure.5. The expression of β-glucuronidase induced by Iptg with the existence of LacI can be measured by the enzyme activity. Higher the total activity can be seen directly by deeper purple color. The activity can also be measured at OD540 with OD600 normalization of bacteria concentration.''' ]] | ||
| + | |||
| + | In the scenario where there is LacI in the plasmid and also where there exists LacO in the upstream gene of β-glucuronidase, this enzyme can be expressed in E. coli BL21 with Iptg induction. The enzymic activities and therefore the catalytic capabilities of the cells and the bacteria culture can therefore be controlled with different concentrations of induction substance. | ||
| + | <br/> | ||
| + | |||
| + | <b>Summary</b> | ||
| + | LacI can effectively work together with LacO to control gene expression in E. coli BL21(DE3). Our results confirmed 0.5 to 2mM Iptg can be effective and sufficient to release the suppression of gene expression exerted by LacI protein. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <br/><br/> | ||
| + | = CKWA-China 2021 Contribution = | ||
| + | |||
| + | We use LacI for expression of Lon protease. The E. coli we used are BL21, Rosetta and similar bateria. According to the reference articles, this Lon protease decomposes C-Myc protein commonly found in cancer cells. Because of this potential, the protease is planned to be used to make medicine to suppress cancer recurrence. | ||
| + | |||
| + | |||
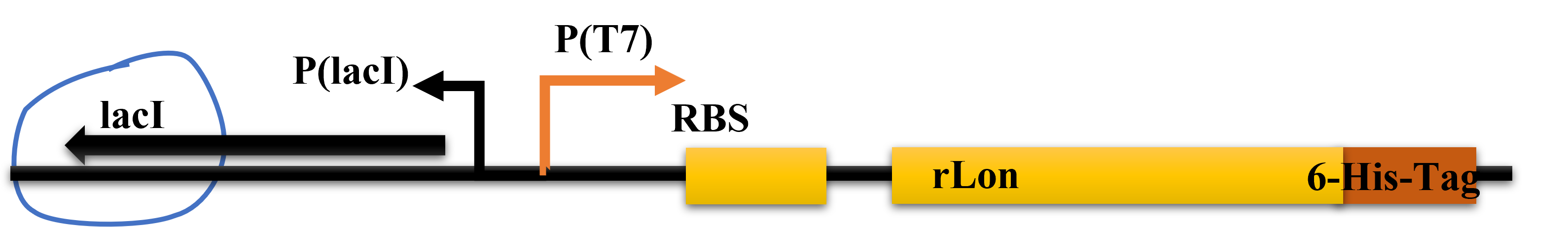
| + | [[File:T--CKWA-China-Lac.png|thumb|left|400px|'''Figure.1.The designed use of the part LacI.]] | ||
| + | |||
| + | We designed to use LacI and LacO together to produce the Lon protease in a commonly used plasmid. The plasmid structure works as shown in the figure below. | ||
| + | |||
| + | For the measurement, E. coli Rosetta containing the designed plasmid was incubated at 37C overnight with 1mM IPTG added. Then the cells are lysed by 200 μL 4% SDS for 10 minutes in room temperature, then 10 min at 95C. Then loading buffer is added. The SDS-PAGE was carried with a 12% precast polyacrylamide gel. Coomassie bright blue stain is used to show the bands. | ||
| + | <br/> | ||
| + | |||
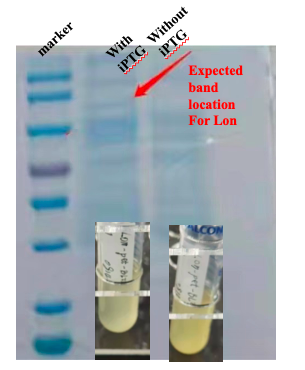
| + | [[File:T--CKWA-China-Lon1.jpg|thumb|left|400px|'''Figure.2. Bacteria Culture overnight and the SDS-PAGE results with/without iPTG.]] | ||
| + | <br/> | ||
| + | With this induction conditions, first test failed to observe a clear band of Lon protein. The bacteria culture was also relatively clear. It is possible that Lon protease has been decomposing the proteins needed for E. coli growth. Because the bacteria growth was suppressed by the Lon protease, its own production was low. | ||
| + | |||
| + | <br/> | ||
| + | |||
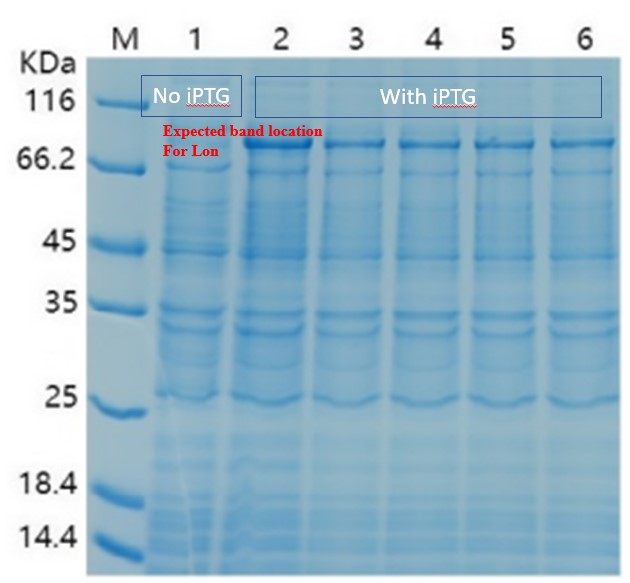
| + | [[File:T--CKWA-China-Lon2.jpg|thumb|left|400px|'''Figure.3. Expression of Lon protease confirmed in SDS-PAGE. ]] | ||
| + | <br/> | ||
| + | |||
| + | When there is LacI and there is LacO before the Lon gene sequence, IPTG can be used to control the expression. The control can be tuned by the concentration and timing of induction. We then changed our condition: the bacteria were cultured on LB plate overnight at 37C, and then pick a single colony and incubate in LB medium until the OD600 reached 0.6-0.8. At this time, IPTG was added at 0.5 mM, and the medium was again incubated at 37C for another 4 hours. This time the SDS-PAGE results showed remarkable expression of the Lon protease. The results confirmed the capability of this part, and also showed for better protein expression and better bateria growth, the timing for induction should also be considered and experimented. | ||
| + | <br/> | ||
| + | |||
| + | '''Summary''' | ||
| + | This part LacI can work with LacO in E. coli Rosseta to control the protein production. The control power has important meanings for some special proteins like Lon protease, which can suppress bacteria growth. Our results suggested at 0.5mM IPTG concentration and an induction timing at OD600=0.6~0.8 can help improve the production target protein. | ||
| + | |||
| + | ==Contribution == | ||
| + | |||
| + | *'''Group:''' [https://2021.igem.org/Team:SCU-China SCU-China iGEM team 2021] | ||
| + | *'''Author:''' Yilong Xu | ||
| + | |||
| + | ===Characterization === | ||
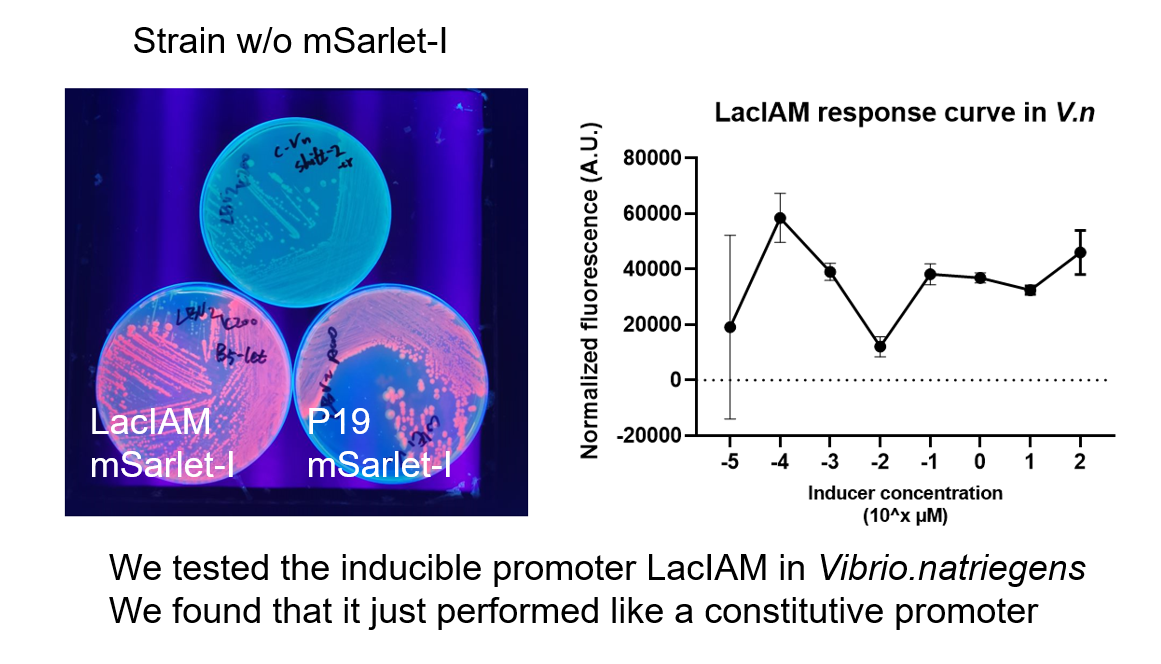
| + | We tested the inducible promoter LacIAM in Vibrio.natriegens, We found that it just performed like a constitutive promoter | ||
| + | |||
| + | [[image: T--SCU-China--scu-2021-LACI.png|600px|center|]] | ||
| + | |||
| + | |||
| + | ==IMPROVEMENT REFERENCE: WHU-China 2022== | ||
| + | In order to maintain the stable period of our oscillator “Repressilator”, we designed the Directed Evolution Module to develop LacI repressor, making it only responsible to high-level lactose that hardly exists in normal intestine environment. Using site-saturation mutagenesis, we created a mutant library and drive the ideal variant through series of selective culture of the engineered bacteria. Our developed LacI repressor showed no expression under low lactose condition, and the leakage problem of the lac promoter is also greatly improved after this mutation. <br> | ||
| + | |||
| + | The new part is: BBa_K4144041(Group: iGEM22_WHU-China), Designed by: Riqui Pi. | ||
Latest revision as of 15:03, 11 October 2022
lacI repressor from E. coli (+LVA)
Coding region for the LacI protein with an LVA degradation tail and without an RBS. LacI binds to the pLac regulator BBa_R0010 and PLlac01 hybrid regulator BBa_R0011 and inhibits transcription. [http://openwetware.org/wiki/IPTG IPTG (Isopropylthiogalactoside)] binds to LacI and inhibits its operation, therefore promoting transcription.
A rapid degradation tail (LVA) has been added to improve the switch time for High to Low performance of this part.
Usage and Biology
This particular LacI protein was derived from E. coli and contributed by Michael Elowitz. (See Part Design for more information.)
- Allergen characterization of BBa_C0012: Not a potential allergen
The Baltimore Biocrew 2017 team discovered that proteins generated through biobrick parts can be evaluated for allergenicity. This information is important to the people using these parts in the lab, as well as when considering using the protein for mass production, or using in the environment. The allergenicity test permits a comparison between the sequences of the biobrick parts and the identified allergen proteins enlisted in a data base.The higher the similarity between the biobricks and the proteins, the more likely the biobrick is allergenic cross-reactive. In the full-length alignments by FASTA, 30% or more amount of similarity signifies that the biobrick has a Precaution Status meaning there is a potential risk with using the part. A 50% or more amount of identity signifies that the biobrick has a Possible Allergen Status. In the sliding window of 80 amino acid segments, greater than 35% signifies similarity to allergens. The percentage of similarity implies the potential of harm biobricks’ potential negative impact to exposed populations. For more information on how to assess your own biobrick part please see the “Allergenicity Testing Protocol” in the following page http://2017.igem.org/Team:Baltimore_Bio-Crew/Experiments
For the biobrick Part:BBa_C0012, there was a 0% of identity match and 0% similarity match to the top allergens in the allergen database. This means that the biobrick part is not of potential allergen status. In 80 amino acid alignments by FASTA window, no matches found that are greater than 35% for this biobrick. This also means that there is not of potential allergen status.
>Internal Priming Screening Characterization of BBa_C0012: Has 1 possible internal priming site between this BioBrick part and the VF2 primer. This BioBrick part also has 4 possible internal priming site between this part and the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
For the BioBrick part BBa_C0012, the first location of the internal priming site is on the 207-214 base number of the BioBrick and on the 1-8 base number of the VF2 primer. The first location of the internal priming site is on the 628-622 base number of the BioBrick and on the 13-19 base number of the VR primer. The second location of the internal priming site is on the 947-941 base number of the BioBrick and on the 4-10 base number of the VR primer. The third location of the internal priming site is on the 778-784 base number of the BioBrick and on the 1-7 base number of the VR primer. The fourth location of the internal priming site is on the 1015-1021 base number of the BioBrick and on the 4-10 base number of the VR primer.
Tsinghua 2018's characterization
The Relation between LacI Concentration and IPTG Induction Efficiency
I Background
In order to investigate how lacI dosage affects IPTG induction, we used Anderson promotor J23100, J23110 and J23114 to design three constitutive lacI generators of different intensities. The three lacI generators were then ligated with Ptac driven reporter sfGFP to make three IPTG induction devices (BBa_K2558203, BBa_K2558204, BBa_K2558205). By measuring sfGFP fluorescence we tested how these devices react to IPTG.
II Results
With high level of lacI expression (BBa_K2558203), sfGFP fluorescence had almost no response to IPTG induction. Weak lacI expression (BBa_K2558205) had the most significant IPTG induced sfGFP expression. With medium lacI expression level (BBa_K2558204), the induction efficiency lay in between. Therefore, the result proves that high level of lacI expression severely decreases IPTG induction efficiency [1]. Furthermore, IPTG concentration can affect the regulation part performance. The figure shows that without IPTG the sfGFP florescence intensity remained low. After IPTG addition, fluorescence signal immediately began to climb, forming a peak at five hours after induction, then sfGFP florescence intensity decreased and maintained at a lower level afterwards. IPTG concentration did not significantly affect the height of the peak or the expression level after the peak, but rather the peak width and expression stability of the system. Figures indicate that 5-10 mM IPTG had the most stable induction results.
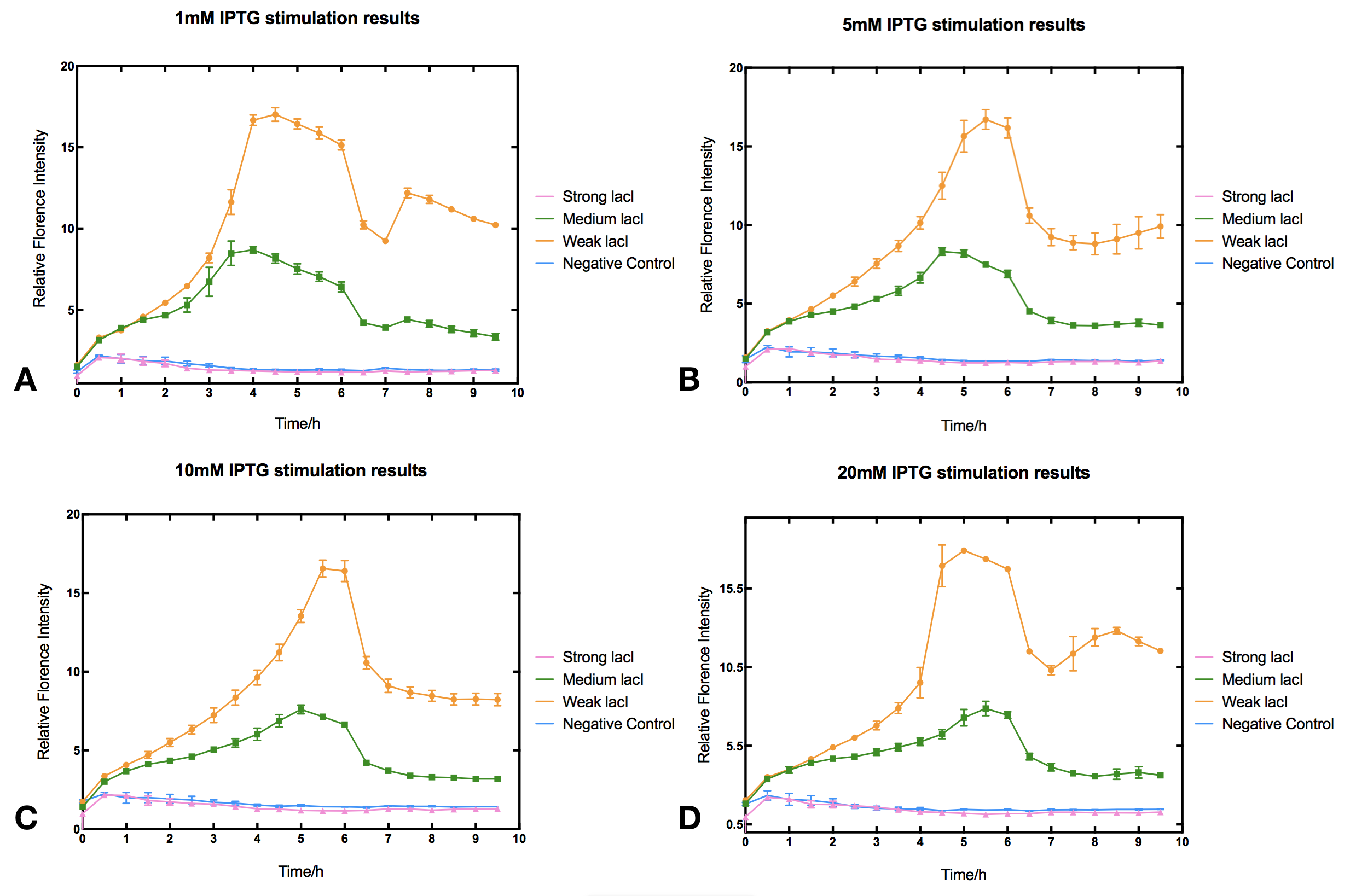
III Protocol
- one Transform the plasmids into E. coli DH5α.
- two Pick a single colony by a sterile tip from each of the LB plates for all the experimental and control groups. Add the colony into 5ml M9 medium with ampicillin at 100 ng/µl. Incubate for 6-8 h at 37℃ in a shaker.
- three Measure OD600 of the culture medium with photometer. Dilute the culture medium until OD600 reaches 0.6.
- four Add 100 µl bacteria culture medium into a sterile 96-well plate. Add IPTG to final concentrations of 0, 1, 5, 10, 20 mM. Fresh M9 medium serves as blank control. Positive control is colony constantly expressing sfGFP and negative control is colony without sfGFP expression. Place the 96-well plate into an automatic microplate reader. Incubate at 16℃ overnight and record the fluorometric value at 510 nm and OD600 for each well every 30 minutes.
- five Each group should be repeated for at least 3 times.
IV Reference
[1] Szabolcs Semsey, Sandeep Krishna. "The effect of LacI autoregulation on the performance of the lactose utilization system in Escherichia coli" Nucleic Acids Res 2013 Jul; 41(13): 6381–6390
2019 Fudan-TSI's Improvement
Overview
This year team Fudan-TSI has upgraded LacI repressor coding gene to a new version (BBa_K3257045 https://parts.igem.org/Part:BBa_K3257045). With lacIq promoter (BBa_K3257003 https://parts.igem.org/Part:BBa_K3257003) and rrnB T1 terminator (BBa_K3257020 https://parts.igem.org/Part:BBa_K3257020), improved LacI protein can be expressed and function properly in the Escherichia coli BL21(DE3). We used EGFP as a reporter controlled by our improved Lac operon and measured its green fluorescence over time.
For detailed design, please refer to https://parts.igem.org/Part:BBa_K3257045.
For a composite LacI repressor protein expression system, please refer to https://parts.igem.org/Part:BBa_K3257100.
According to our experiment, our Lac operon is improved in the following three main aspects.
Results
Lower Response to Arabinose, Better Orthogonality
Cross talk between the response to IPTG and arabinose has been a defect of the wild type Lac operon. When 4mM arabinose added, a few lac inhibitors detach from lac operator which means that it is induced in a relatively low but unignorable level. According to the measurement of our experiment, our improved LacI can respond to IPTG with better orthogonality. The figure below (Figure 1) is the measurement of the fluorescence of EGFP controlled by wild-type and improved Lac operon. It shows that when 4mM arabinose is added, oLacI(wild-type LacI) is induced at a significantly high level while iLacI(improved LacI) is induced at a lower level.
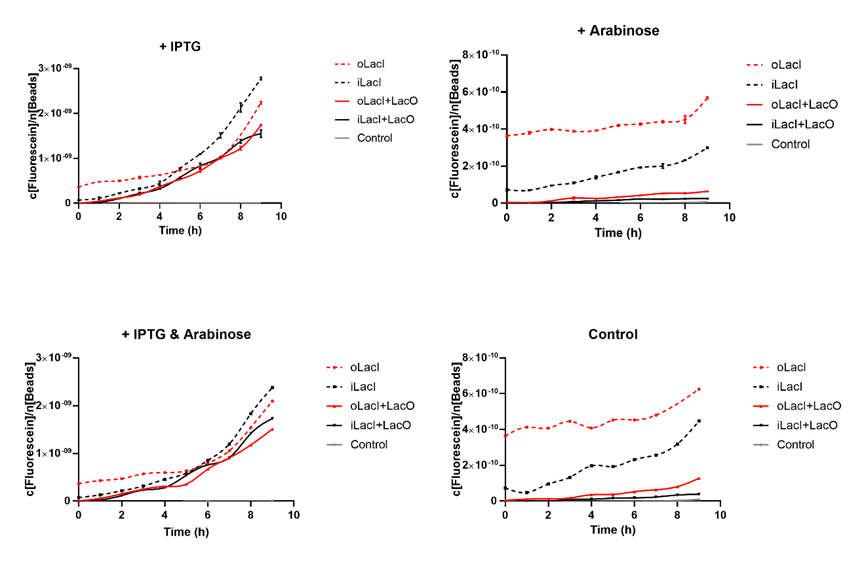
Higher level of induction than the Wild-type Lac operon when Induced by IPTG
Next step we prove that our improved Lac operon can be better induced by IPTG, which counts a great deal for the usage of this operon. The figure below (Figure 2) is the measurement of the fluorescence of EGFP controlled by wild-type and improved Lac operon. It shows that when 1mM IPTG is added, EGFP controlled by both operons can be disinhibited and the improved LacI is induced at a relatively higher level.

Lower Uninduced Leakage
The uninduced leakage level is also an important parameter of an operon. Improved LacI lowers the leakage level compared to the wild-type one. The figure below (Figure 3) is the measurement of the fluorescence of EGFP controlled by wild-type and improved Lac operon. When no IPTG or arabinose is added, the fluorescence of EGFP controlled by improved Lac operon is under the detection range while the fluorescence of EGFP controlled by wild-type Lac operon remains a detectable signal indicating a considerably undesired leakage.
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
References
[1] Adam J. Meyer, Thomas H. Segall-Shapiro, Emerson Glassey, Jing Zhang & Christopher A. Voigt. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nature Chemical Biology volume 15, pages196–204 (2019)
BHSF 2020's characterization
This operator can not only be used in prokaryotic promoters, but also in eukaryotic promoters when inserted downstream of TATA-box.
After we decided to use toggle switch as the principle of our circuit, and use lac as a molecule in our circuit, we used these keywords to find the most suitable system for our yeast, and we found the article describing a hybrid promoter:
1.If a lac operator site (lacO) is positioned appropriately downstream of the TATA‐box, gene expression from a nmt::lacO promoter (hybrid promoter) can be greatly repressed by lacI inhibitor.
2.Insertion of the operator sequence into the nmt promoter 5 bp downstream of TATA-box causes irreversible repression of gene expression when lacI is present.
3.When the operator in inserted directly downstream of TATA-box, the addition of IPTG to the medium could relieve the repression to some extent, but it is not fully reversible.
References
Kjaerulff S, Nielsen O. An IPTG-inducible derivative of the fission yeast nmt promoter. Yeast. 2015 Jun;32(6):469-78. doi: 10.1002/yea.3073. Epub 2015 Mar 31. Erratum in: Yeast. 2016 Jan;33(1):33. PMID: 25801050.
iBowu-China 2021's characterization
I Design
II Introduction
This year team iBowu-China used two parts LacI and LacO, with iPTG as induction methods, to control the expression of sfGFP protein and β-glucuronidase protein in E. coli BL21(DE3)
III Protocol
- Transform the plasmids into E. coli BL21(DE3)
- Pick a single colony by a sterile tip from each of the LB plates for all the experimental and control groups. Add the colony into 4ml LB medium with kanamycin. Add 1mM iPTG to all experimental groups. Incubate at 37℃ in a shaker overnight.
- Add 100 µl bacteria culture medium into a sterile 96-well plate. Measure fluorescence at 488nm excitation and 502nm absorption, and then also OD600 for normalization, with a microplate reader.
IV Results
sfGFP
Under the condition that there exists the part LacI, and there also exists LacO in the upstream gene of sfGFP, Iptg induction can control the switch on and off of the expression of green fluorescence protein sfGFP.
The plot and photo demonstrate that 0.5nM Iptg added into the medium is sufficient to induce the expression of sfGFP with the existence of lacI part.
Enzyme activity of β-glucuronidase
In the scenario where there is LacI in the plasmid and also where there exists LacO in the upstream gene of β-glucuronidase, this enzyme can be expressed in E. coli BL21 with Iptg induction. The enzymic activities and therefore the catalytic capabilities of the cells and the bacteria culture can therefore be controlled with different concentrations of induction substance.
Summary LacI can effectively work together with LacO to control gene expression in E. coli BL21(DE3). Our results confirmed 0.5 to 2mM Iptg can be effective and sufficient to release the suppression of gene expression exerted by LacI protein.
CKWA-China 2021 Contribution
We use LacI for expression of Lon protease. The E. coli we used are BL21, Rosetta and similar bateria. According to the reference articles, this Lon protease decomposes C-Myc protein commonly found in cancer cells. Because of this potential, the protease is planned to be used to make medicine to suppress cancer recurrence.
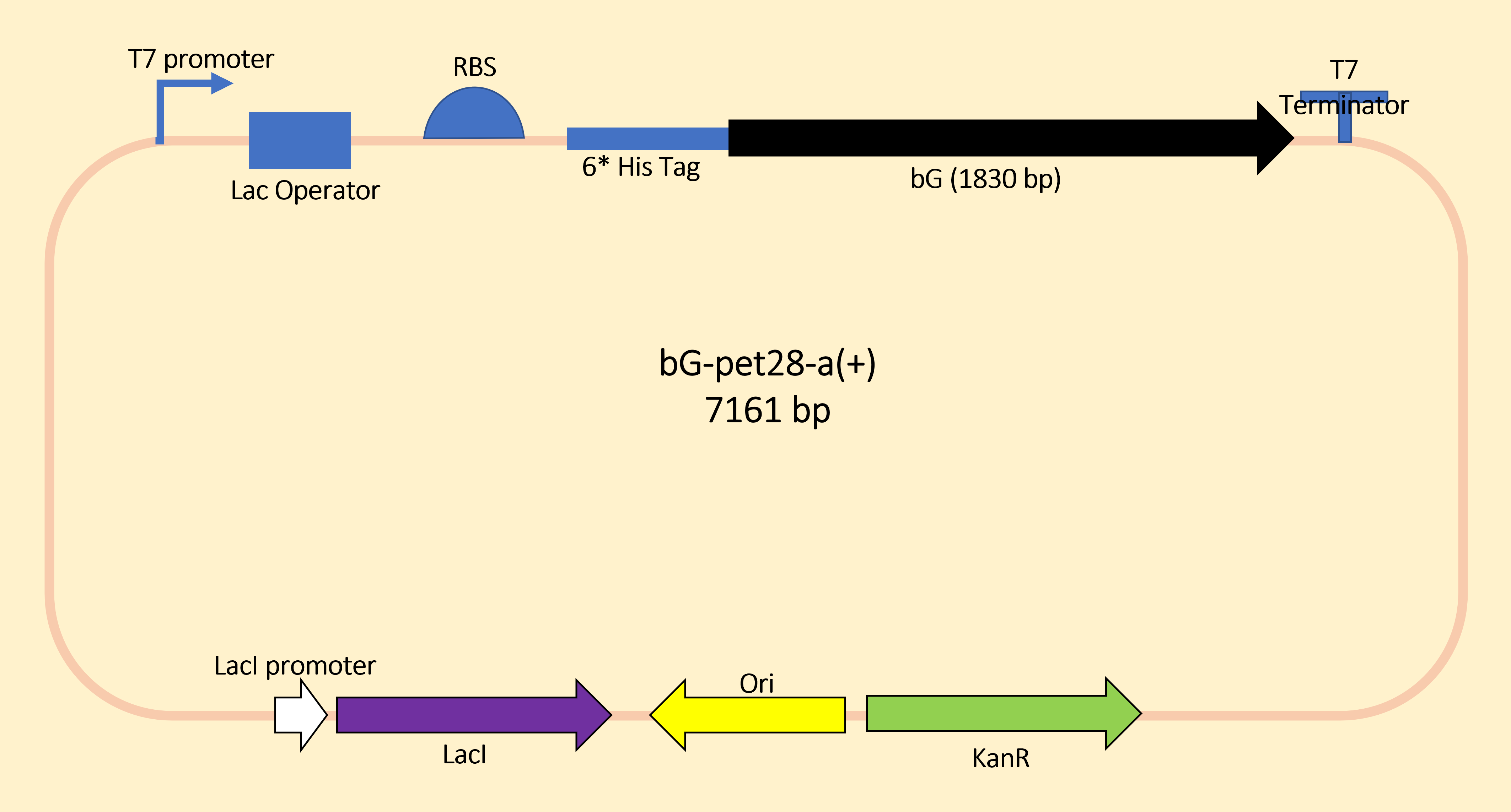
We designed to use LacI and LacO together to produce the Lon protease in a commonly used plasmid. The plasmid structure works as shown in the figure below.
For the measurement, E. coli Rosetta containing the designed plasmid was incubated at 37C overnight with 1mM IPTG added. Then the cells are lysed by 200 μL 4% SDS for 10 minutes in room temperature, then 10 min at 95C. Then loading buffer is added. The SDS-PAGE was carried with a 12% precast polyacrylamide gel. Coomassie bright blue stain is used to show the bands.
With this induction conditions, first test failed to observe a clear band of Lon protein. The bacteria culture was also relatively clear. It is possible that Lon protease has been decomposing the proteins needed for E. coli growth. Because the bacteria growth was suppressed by the Lon protease, its own production was low.
When there is LacI and there is LacO before the Lon gene sequence, IPTG can be used to control the expression. The control can be tuned by the concentration and timing of induction. We then changed our condition: the bacteria were cultured on LB plate overnight at 37C, and then pick a single colony and incubate in LB medium until the OD600 reached 0.6-0.8. At this time, IPTG was added at 0.5 mM, and the medium was again incubated at 37C for another 4 hours. This time the SDS-PAGE results showed remarkable expression of the Lon protease. The results confirmed the capability of this part, and also showed for better protein expression and better bateria growth, the timing for induction should also be considered and experimented.
Summary This part LacI can work with LacO in E. coli Rosseta to control the protein production. The control power has important meanings for some special proteins like Lon protease, which can suppress bacteria growth. Our results suggested at 0.5mM IPTG concentration and an induction timing at OD600=0.6~0.8 can help improve the production target protein.
Contribution
- Group: SCU-China iGEM team 2021
- Author: Yilong Xu
Characterization
We tested the inducible promoter LacIAM in Vibrio.natriegens, We found that it just performed like a constitutive promoter
IMPROVEMENT REFERENCE: WHU-China 2022
In order to maintain the stable period of our oscillator “Repressilator”, we designed the Directed Evolution Module to develop LacI repressor, making it only responsible to high-level lactose that hardly exists in normal intestine environment. Using site-saturation mutagenesis, we created a mutant library and drive the ideal variant through series of selective culture of the engineered bacteria. Our developed LacI repressor showed no expression under low lactose condition, and the leakage problem of the lac promoter is also greatly improved after this mutation.