Part:BBa_K3257100
Improved LacI Expression System
This part is the system we build to express our improved LacI. With lacIq promoter and rrnB T1 terminator, improved LacI protein can be expressed and function properly in the Escherichia coli BL21(DE3). We used EGFP as a reporter controlled by our improved Lac operon and measured its green fluorescence over time.
According to our experiment, our Lac operon is improved in the following three main aspects.
Lower Response to Arabinose, Better Orthogonality
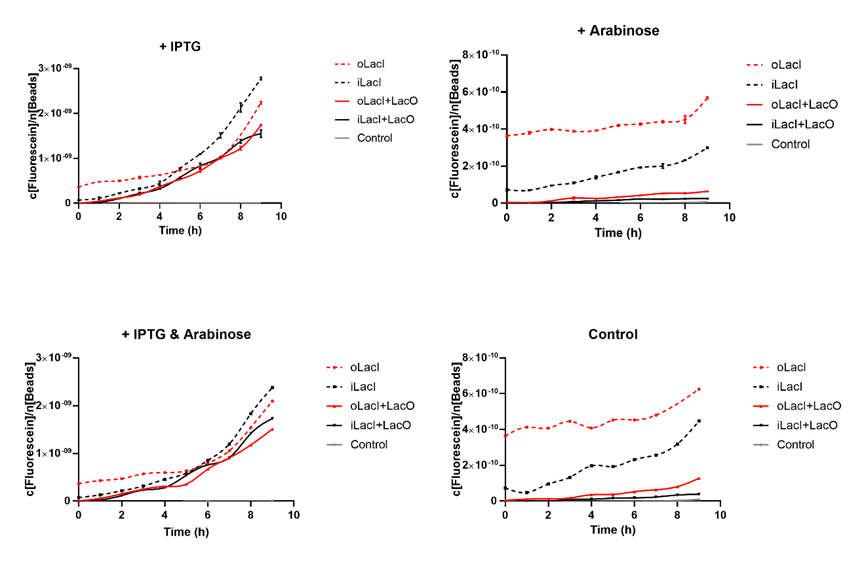
Cross talk between the response to IPTG and arabinose has been a defect of the wild type Lac operon. When 4mM arabinose added, a few lac inhibitors detach from lac operator which means that it is induced in a relatively low but unignorable level. According to the measurement of our experiment, our improved LacI can respond to IPTG with better orthogonality. The figure below (Figure 1) is the measurement of the fluorescence of EGFP controlled by wild-type and improved Lac operon. It shows that when 4mM arabinose is added, oLacI(wild-type LacI) is induced at a significantly high level while iLacI(improved LacI) is induced at a lower level.
Higher Induction Level than the Wild-type Lac operon when Induced by IPTG
Next step we prove that our improved Lac operon can be normally induced by IPTG, which counts a great deal for the usage of this operon. The figure below (Figure 2) is the measurement of the fluorescence of EGFP controlled by wild-type and improved Lac operon. It shows that when 1mM IPTG is added, EGFP is better induced when it is controlled by the improved Lac operon.

Lower Uninduced Leakage
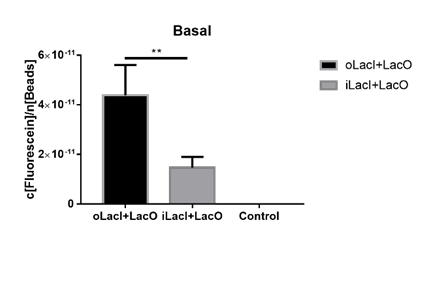
The uninduced leakage level is also an important parameter of an operon. Improved LacI lowers the leakage level compared to the wild-type one. The figure below (Figure 3) is the measurement of the fluorescence of EGFP controlled by wild-type and improved Lac operon. When no IPTG or arabinose is added, the fluorescence of EGFP controlled by improved Lac operon is under the detection range while the fluorescence of EGFP controlled by wild-type Lac operon remains a detectable signal indicating a considerably undesired leakage.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal XbaI site found at 1258
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal XbaI site found at 1258
- 25INCOMPATIBLE WITH RFC[25]Illegal XbaI site found at 1258
- 1000COMPATIBLE WITH RFC[1000]
Reference
AJ Meyer et al. Escherichia coli "Marionette" strains with 12 highly optimized small-molecule sensors. Nat Chem Biol. 2019 Feb;15(2):196-204. doi: 10.1038/s41589-018-0168-3. Epub 2018 Nov 26.
| None |
