Help:Ribosome Binding Site
![]() Browse Ribosome Binding Site parts!
Browse Ribosome Binding Site parts!
A ribosome binding site (RBS) is a segment of the 5' (upstream) part of an mRNA molecule that binds to the ribosome to position the message correctly for the initiation of translation.
The RBS controls the accuracy and efficiency with which the translation of mRNA begins.
Contents
Biology and background
In prokaryotes (such as E. coli) the RBS typically lies about 7 nucleotides upstream from the start codon (i.e., the first AUG). The sequence itself in general is called the [http://en.wikipedia.org/wiki/Shine-dalgarno_sequence "Shine-Dalgarno"] sequence after its discoverers, regardless of the exact identity of the bases. Strong Shine-Dalgarno sequences are rich in purines (A's,G's), and the "Shine-Dalgarno consensus" sequence -- derived statistically from lining up many well-characterized strong ribosome binding sites -- has the sequence AGGAGG. The complementary sequence (CCUCCU) occurs at the 3'-end of the structural RNA ("16S") of the small ribosomal subunit and it base-pairs with the Shine-Dalgarno sequence in the mRNA to facilitate proper initiation of protein synthesis.
Protein synthesis in eukaryotes differs from this model. The 5' end of the mRNA has a modified chemical structure ("cap") recognized by the ribosome, which then binds the mRNA and moves along it ("scans") until it finds the first AUG codon. A characteristic pattern of bases (called a "Kozak sequence") is sometimes found around that codon and assists in positioning the mRNA correctly in a manner reminiscent of the Shine-Dalgarno sequence, but not involving base pairing with the ribosomal RNA.
Engineering RBS issues
Efficiency
Different RBSs bind ribosomes with different efficiencies. While the overall efficiency of translation may not be directly proportional to this RBS-binding efficiency, it is an important variable. In the future, we expect that this data book will have a table such as the following:
Standard BioBrick Ribosome Binding Sites
| Part Number | Binding Efficiency | Sequence |
|---|---|---|
| BBa_B0030 | 0.6 | att aaa gag gag aaa |
| BBa_B0031 | 0.07 | tca cac agg aaa cc |
| BBa_B0032 | 0.3 | tca cac agg aaa g |
A BioBrick engineer, armed with this table, could perform a first-order re-engineering of the transfer function by measuring a component with BBa_B0031 and then selecting a more appropriate RBS.
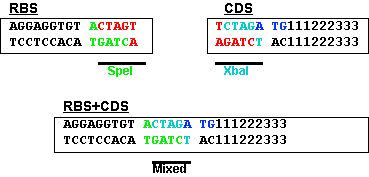
In order to allow for easy variation in the RBS, some BioBrick parts have their RBSs and their protein coding regions implemented as separate parts. For convenience, the combined, normalized part is also available. In order to divide the BioBrick part into its RBS and coding region, a set of BioBrick restriction sites must be designed.
Standard Assembly and the RBS
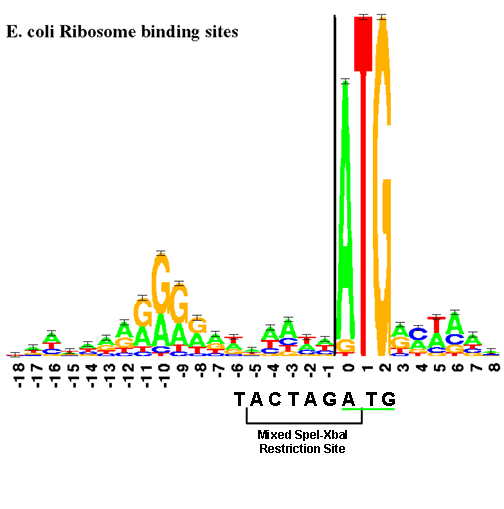
The RBS is physically near the start codon. (In BioBricks, this will always be ATG.) The "sequence logo", similar to a consensus sequence, for the RBS is shown below. The size of each letter is proportional to the frequency of that base in the RBS part.
This figure shows the RBS as an A/G-rich region about 10 bases upstream of the start codon. This leaves little room for a BioBrick restriction enzyme site. The standard assembly method of BioBricks normally results in an 8-base region containing a 6-base mixed SpeI/XbaI restriction site surrounded by a base on each end as shown below. If a start codon followed this sequence, the RBS would be significantly disturbed. see you in versaille
--T ACTAGA G--
This problem is solved by changing the rightmost base from a G to a T. This results in the sequence shown below and limits the impact of the BioBrick overhead to six bases.