Part:BBa_K1033914
amajLime, yellow-green chromoprotein (incl RBS & J23110)
This chromoprotein from the coral Anemonia majano, amajLime (also known as amajCFP or amFP486), naturally exhibits strong color when expressed. The protein has an absorption maximum at 458 nm giving it a yellow-green color visible to the naked eye. Compared to many other chromoproteins, such as amilCP (BBa_K592009), amilGFP (BBa_K592010), spisPink (BBa_K1033932), asPink (BBa_K1033933) and aeBlue (BBa_K864401), the color development is slower. The color is readily observed in both LB or on agar plates after 24-48 hours of incubation. The protein amajLime has significant sequence homologies with proteins in the GFP family.
Characterisation
Team: Humboldt_Berlin 2019
ExPASy ProtParam Results:
Number of amino acids: 228
Molecular weight: 25185.49 Da
Theoretical pI: 7.27
Total number of negatively charged residues (Asp + Glu): 22
Total number of positively charged residues (Arg + Lys): 22
Extinction coefficients:
Ext. coefficient 26150 M-1 cm-1
Abs 0.1% (=1 g/l) 1.038, assuming all pairs of Cys residues form cystines
Ext. coefficient 25900 M-1 cm-1
Abs 0.1% (=1 g/l) 1.028, assuming all Cys residues are reduced
Instability index: The instability index (II) is computed to be 27.07. This classifies the protein as stable.
Aliphatic index: 51.71
Grand average of hydropathicity (GRAVY): -0.379
In order to measure absorbance and fluorescence spectra of amajLime we transformed the construct (consisting of BBa_J23110 Promotor, BBa_B0034 RBS and amajLime coding sequence) into E. coli (DH10B Competent Cells). After cultivation and we lysed the harvested cells according to this protocol.
The fluorescence and absorbance spectra were measured for 24 samples of 150 µl lysate in 96-well plate on TECAN Plate Reader Infinite 200 Pro. In figure 1 you can see the relative absorbance spectrum with a peak at 454 nm (compared to an excitation maximum of 458 nm in the literature [4])including the respective standard deviation. Figure 2 shows the fluorescence spectrum with a peak at 493 nm (compared to an excitation maximum of 486 nm in the literature [4]) including the respective standard deviation. In figure 3 the joined relative absorbance and fluorescence spectra are shown.
| Table 1. Parameters utilized for absorbance and fluorescence spectra | |||
| Parameter | Value | ||
| Number of Samples | 24 | ||
| Wavelength Step Size | 2 | ||
| Absorbance Scan: Excitation Wavelength Measurement Range (nm) | [300-800] | ||
| Fluorescence Scan: Emission Wavelength Measurement Range (nm) | [475-550] | ||
| Fluorescence Scan: Excitation Wavelength (nm) | 445 | ||
| Gain (G) | 52 | ||
| Number of Flashes | 25 | ||
| Integration Time (µs) | 20 | ||
| Lag Time (µs) | 0 | ||
| Settle Time (ms) | 0 | ||
Contribution
Group: Linkoping_Sweden iGEM 2021
Author: Ewelina Bladh
Summary:
In this contribution, we recorded color development in colonies of E.coli cells of strain XL1. In addition to studying the color development, we confirmed amajLime's absorbance maximum wavelength as well as reconfirmed the fluorescence of amajLime. Additionally, the experiments performed show reproducibility of results.
Color Development
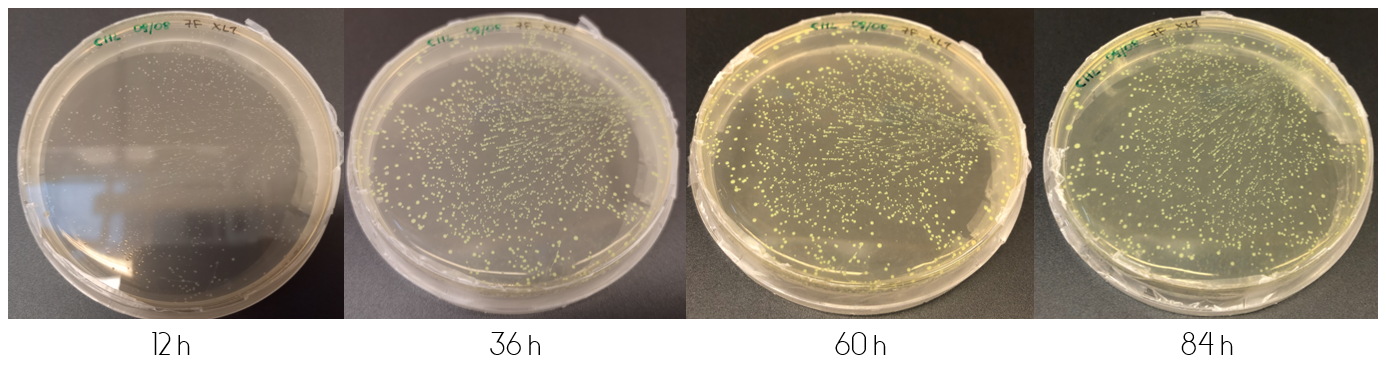
In order to determine the progression of color development in colonies, E.coli cells (XL1) were plated onto LB-Miller agar plates containing a concentration of 25 ug/mL chloramphenicol. The cells were plated in two ways after transformation. The first by plating 100 µL immediately from the liquid culture. The second by first centrifuging the culture, discarding 90% of the supernatant and resuspending the cell pellet in the remaining 10%, followed by plating 100 µL of the resuspension. This results in what is from now on referred to as the diluted and concentrated plate, respectively.
These plates were incubated in 37°C for the following four days, where they were checked upon each day. Figure 1, displayed below, shows the diluted and concentrated plates during this four day study. The illustration of the diluted plate shows that after 12 hours, no color can be seen to the naked eye. This changes over the course of the next 24 hours, where at 36 hours the colonies are clearly visible. The color intensity increases over the next two days. The same can be said about the concentrated plate, but unlike the diluted plate the concentrated plate shows indications of colored colonies already at 12 hours.
Absorbance and Fluorescence
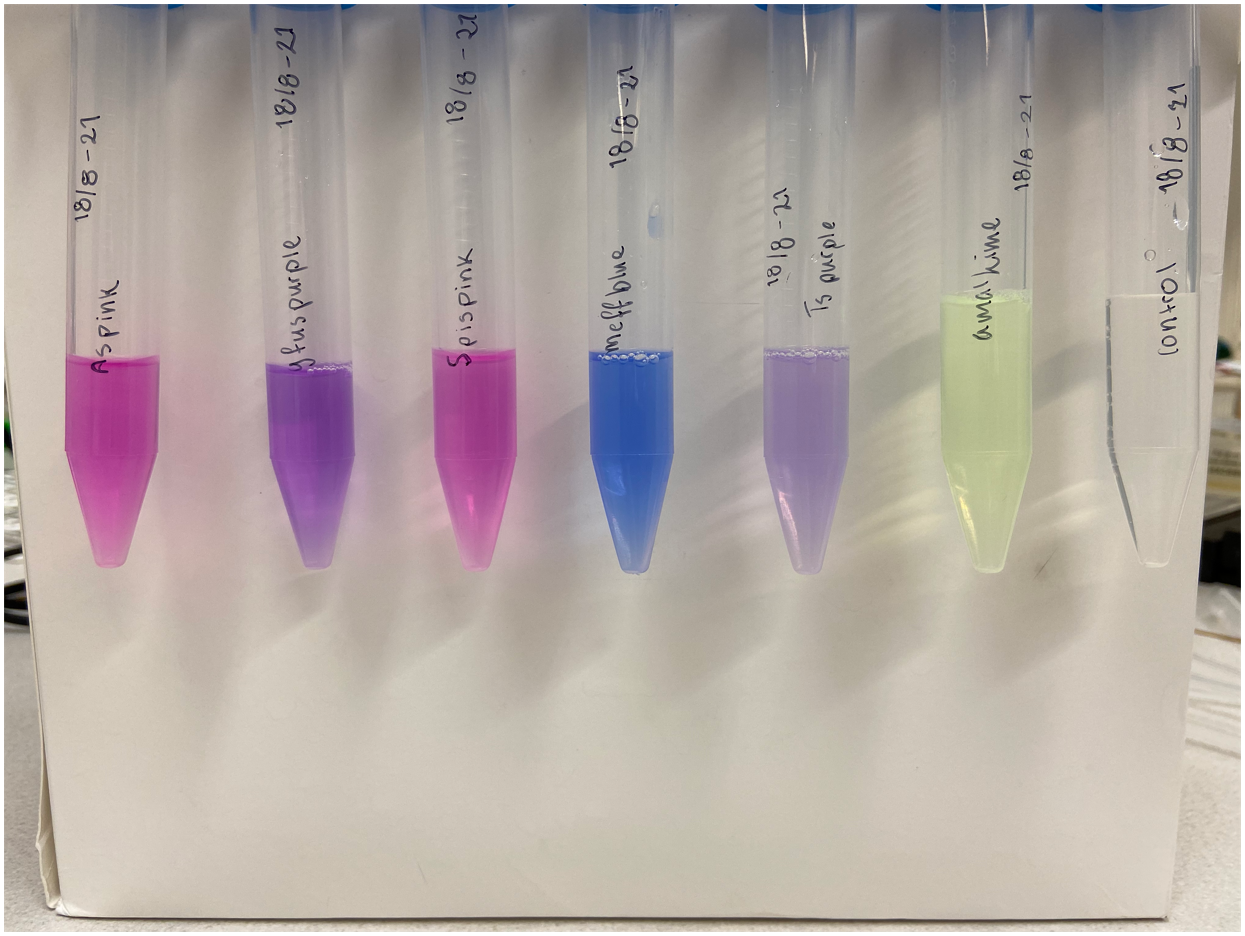
Single colonies displaying a yellow-green color were picked from the diluted plates to create liquid cultures. The liquid cultures consist of 5 mL LB-Miller media and a concentration of 25 µg/mL chloramphenicol. The liquid cultures were incubated at 37°C and 200 rpm for the next two days to ensure protein expression. After incubation, the liquid cultures were centrifuged until a pellet formed. The supernatant was discarded, and the remaining pellet was placed in a -20°C freezer. Cell lysis was done using both water and sonication. The frozen pellet was thawed and 5 mL autoclaved dH2O was added, followed by vortex treatment to resuspend the pellet. Sonication on ice was the next step taken to lyse any cells that did not lyse upon the addition of water. The amplitude of the sonicator was set to 30%, 30 seconds pulsing followed by 30 seconds rest for a total of 2 minutes. Following sonication, the cultures were centrifuged until a pellet formed whereupon the supernatant was transferred to a clean 15 mL Falcon tube and the pellet was discarded. The supernatant of amajLime can be see to the left in Figure 2. Simultaneous experiments with other chromoproteins was also made, their supernatant can be seen with amajLime to the right in Figure 2. The following chromoproteins were also assessed in the same way as amajLime: tsPurple (BBa_K1033903), meffBlue (BBa_K1033900), spisPink (BBa_K1033923), gfasPurple (BBa_K1033917), and asPink (BBa_K1033926).
As a control, plates were also made with untransformed XL1-cells. A colony was picked from the diluted plate to start a liquid culture, followed by lysis like the cells transformed with amajLime. Treatment of control and experimental cells was identical apart from the control culture not being exposed to antibiotics. The supernatant of the control can be seen to the right in Figure 2 along with other chromoproteins.
In order to measure the absorbance, 100 µL supernatant of amajLime and 100 µL supernatant of the control were pipetted into a 96-well plate and measurements were made with BMG Clariostar plate reader. Measurements of absorbance were taken every 2 nm in the range 300 nm to 850 nm. This interval was chosen to include the entire visual spectra including margins. The data yielded from the measurements were subsequently baselined by subtracting the control's absorbance values from amajLime's absorbance values followed by normalization. The resulting graph for amajLime's absorbance can be seen in Figure 3 below. The absorbance maximum of amajLime proved to occur at approximately 454 nm. This result coincides with the results from the team of Humboldt_Berlin 2019 (see above).
When examining the supernatant extracted from amajLime-expressing cells on a UV-table, fluorescence was detected as expected from previous knowledge available on this page. The lysate from the control functioned as a negative control. During the last two days of the color development study, the plates were placed on a UV-table to investigate the fluorescence. As can be seen in Figure 4 below, the colonies fluoresced readily in a turquoise hue. No major difference appear between 60 hours and 84 hours, except from a slight increase in intensity.
References
[http://www.nature.com/nbt/journal/v17/n10/full/nbt1099_969.html] Matz, Mikhail V., et al. "Fluorescent proteins from nonbioluminescent Anthozoa species." Nature biotechnology 17.10 (1999): 969-973.
[http://www.pnas.org/content/102/36/12712.short] Henderson, J. Nathan, and S. James Remington. "Crystal structures and mutational analysis of amFP486, a cyan fluorescent protein from Anemonia majano." Proceedings of the National Academy of Sciences of the United States of America 102.36 (2005): 12712-12717.
[http://www.ncbi.nlm.nih.gov/pubmed/18648549] Alieva, N. O., et al. 2008. Diversity and evolution of coral fluorescent proteins. PLoS One 3:e2680.
[1] Liljeruhm, Josefine et al. “Engineering a palette of eukaryotic chromoproteins for bacterial synthetic biology.” Journal of biological engineering vol. 12:8. 10 May. 2018, doi:10.1186/s13036-018-0100-0
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//function/reporter/color
//function/reporter/fluorescence
| emission | 486 nm |
| excitation | 450 nm |