Part:BBa_B0034
RBS (Elowitz 1999) -- defines RBS efficiency
RBS based on Elowitz repressilator.
Usage and Biology
IIT Madras 2016's Characterization
Modelling
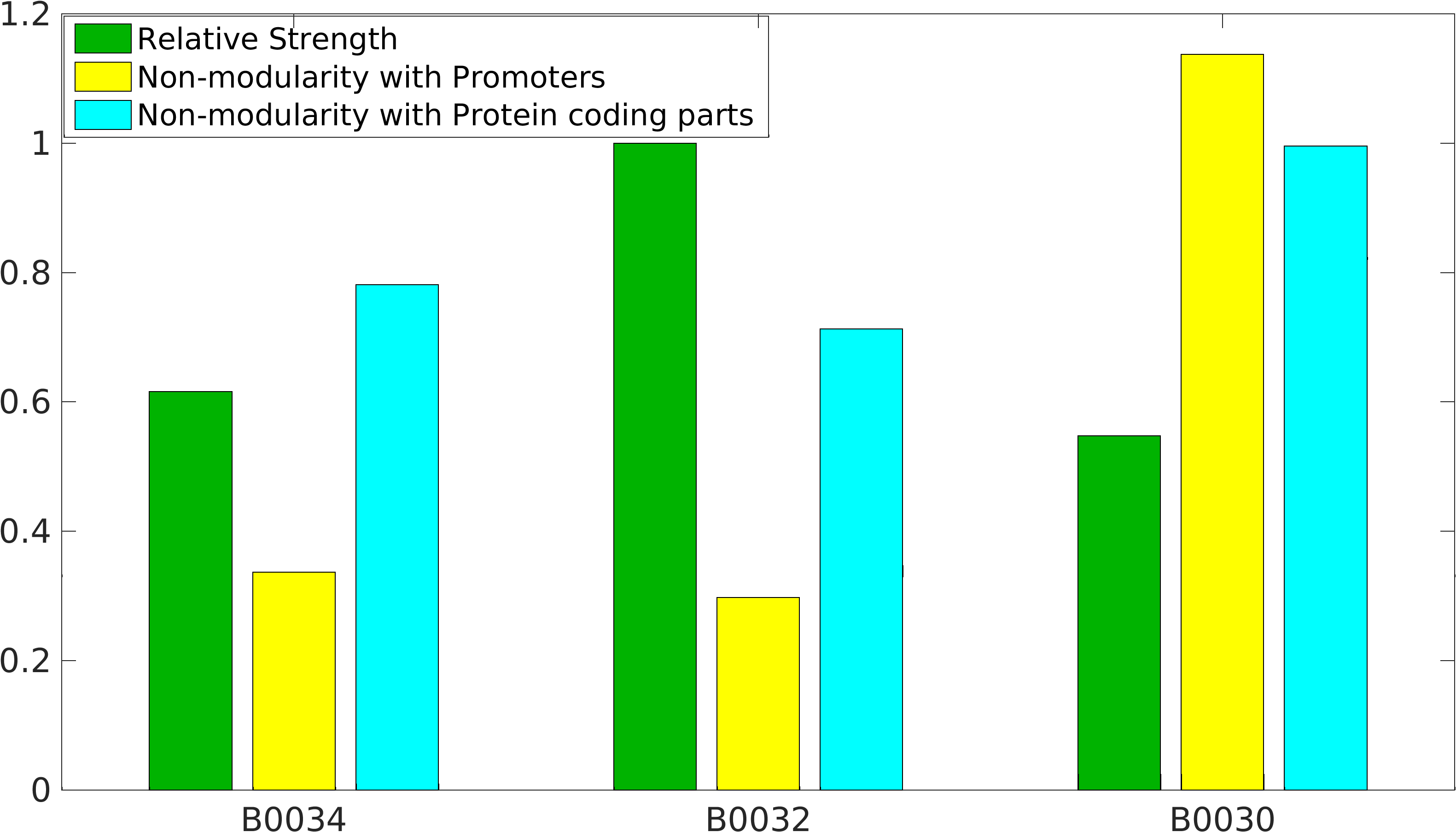
Global non-modularity towards promoters & protein coding parts and relative strength was estimated for RBSs B0030, B0032, B0034 in our [http://2016.igem.org/Team:IIT-Madras/Model#Modularity_of_RBS_parts modelling]
Experimentation
Biobrick RBSs B0030, B0031, B0032, B0034 were used in our 'Noise in Device' experiment to understand the role of RBS parts in giving rise to noise.
Note: The Elowitz RBS is the definition of efficiency 1.0.
Team Warsaw 2010's measurement
definition of 100% strength in our measurement.KEYSTONE_2022 Characterization
As a biobrick RBS (Ribosome-Binding Site), we used it to be assembled with several CDS, such as ClSS_S533A (Part Number: BBa_K4274000) ,CmR29M2-ClSS_S533A (Part Number:BBa_K4274001),ERG20 (Part Number: BBa_K849001), ERG20_F96W(Part Number: BBa_K4274002).
Santalene production in E.coli DH5α
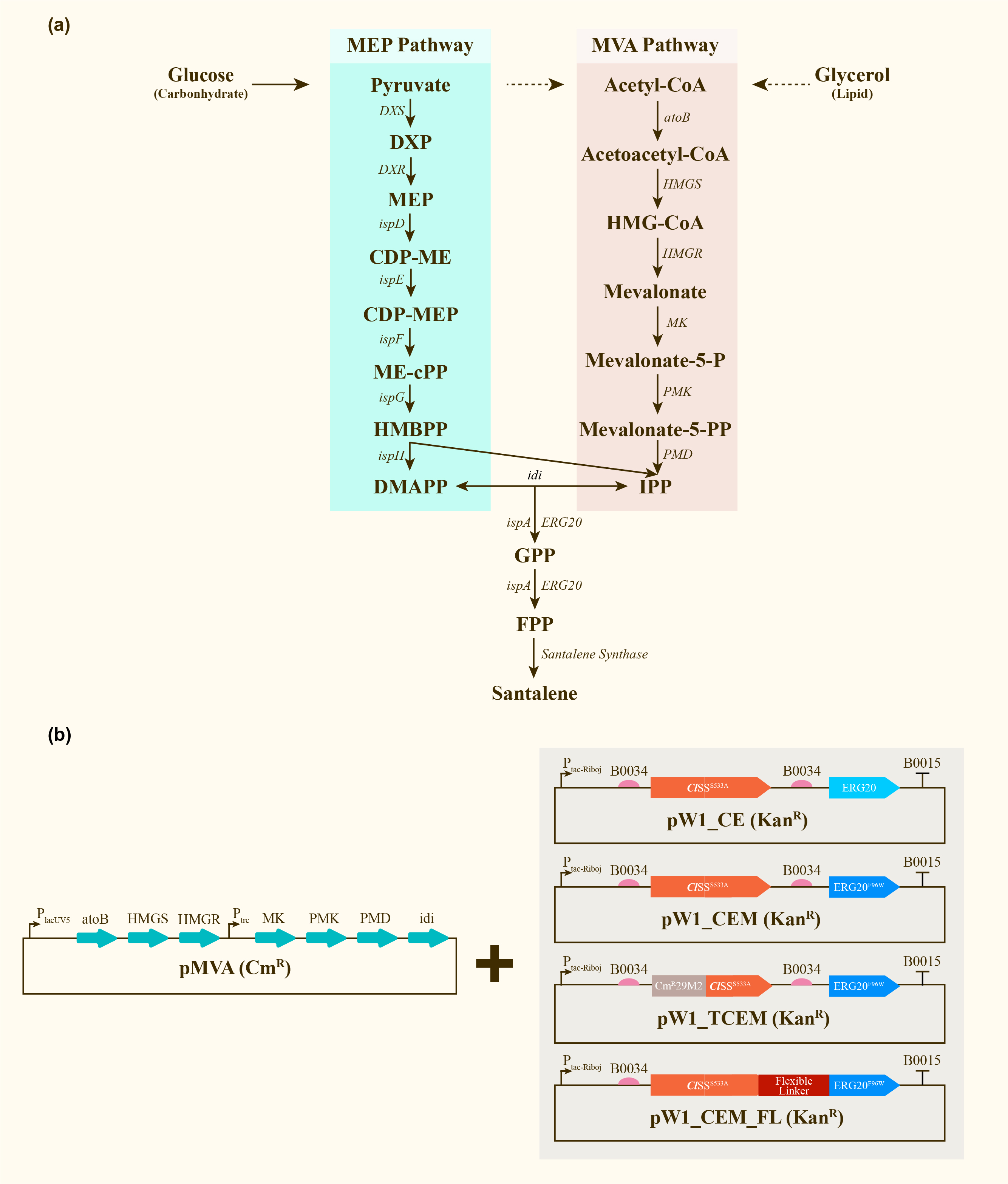
After engineering, E. coli could utilize both MEP pathway and MVA pathway for the universal precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), then synthesize santalene with the help of FPP Synthase (FPPS) and santalene synthase (SS). Except heterologously expressed MVA pathway and ERG20 of Saccharomyces cerevisiae and santalene synthase of Clausena lansium (ClSS), several modifications upon ERG20 or ClSS by amino acid mutation, binding to a hydrophillic tag and the construction of fusion protein were tested for the higher yield of santalene. Therefore, with the help of the co-transformation of pMVA plasmid with various pW1 plasmids, including pW1_CE, pW1_CEM, pW1_TCEM and pW1_CEM_FL, different strains like CE, CEM, TCEM, CEM_FL were successfully constructed (Figure 1). The complete pathway we designed for producing santalene in E. coli is illustrated in Figure 1.
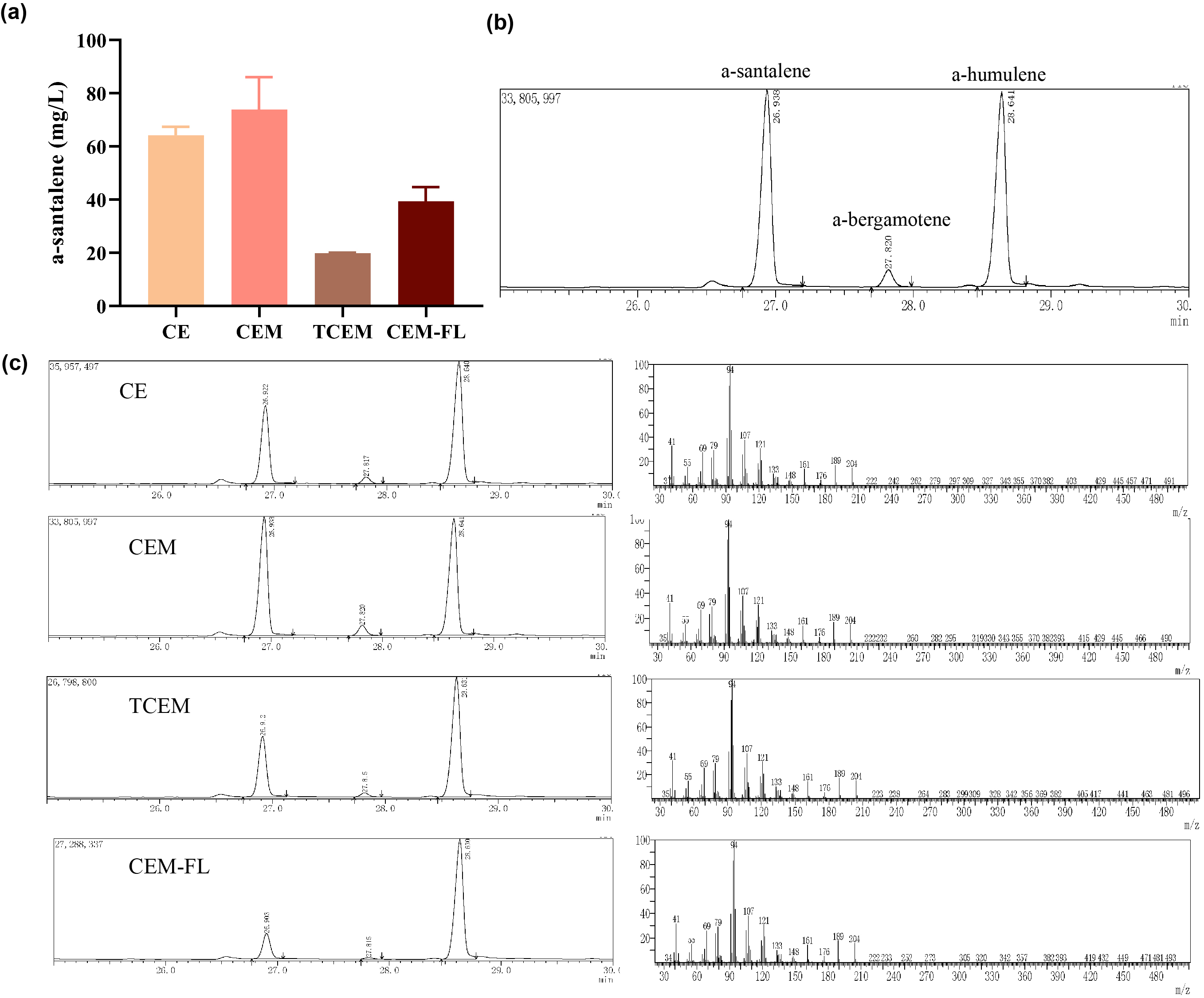
Afterwards, the various engineering of E.coli DH5α ∆TnaA mentioned above were used for santalene production. After rapid centrifugation, the supernatant of dodecane was spiked with with 0.475 g/L a-humulene as an internal standard, and then injected into GC/MS for verification of α-santalene production. It turned out that all samples from four strains appeared a significant peak at the retention time of 26-27 min, and various peak area of different samples exhibited santalene production with differing levels, indicating the general success of E. coli engineering. It can be concluded that the E. coli strain CEM (with pW1_CEM plasmid) produces the maximal level of α-santalene compared to other strains (73.93 mg/L). Furthermore, our study elucidates that the mutation of 96th amino acid into tryptophan could increase the yield of α-santalene by about 20%, substantiating the prominent performance of ERG20F96W in enhancing the supply of FPP and α-santalene production in E. coli (Figure 2).
Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We have adapted the part to be able to assemble transcriptional units with the Golden Gate method and we have done the characterization of this RBS.
Documentation:
BBa_K2656011 is the RBS B0034 standardized into the Golden Braid assembly method. It also includes the BioBrick equivalent scar in the 3' extreme, so the insertion of this supplementary bases ensure correct spacing for the CDS expression when assembled into a TU.
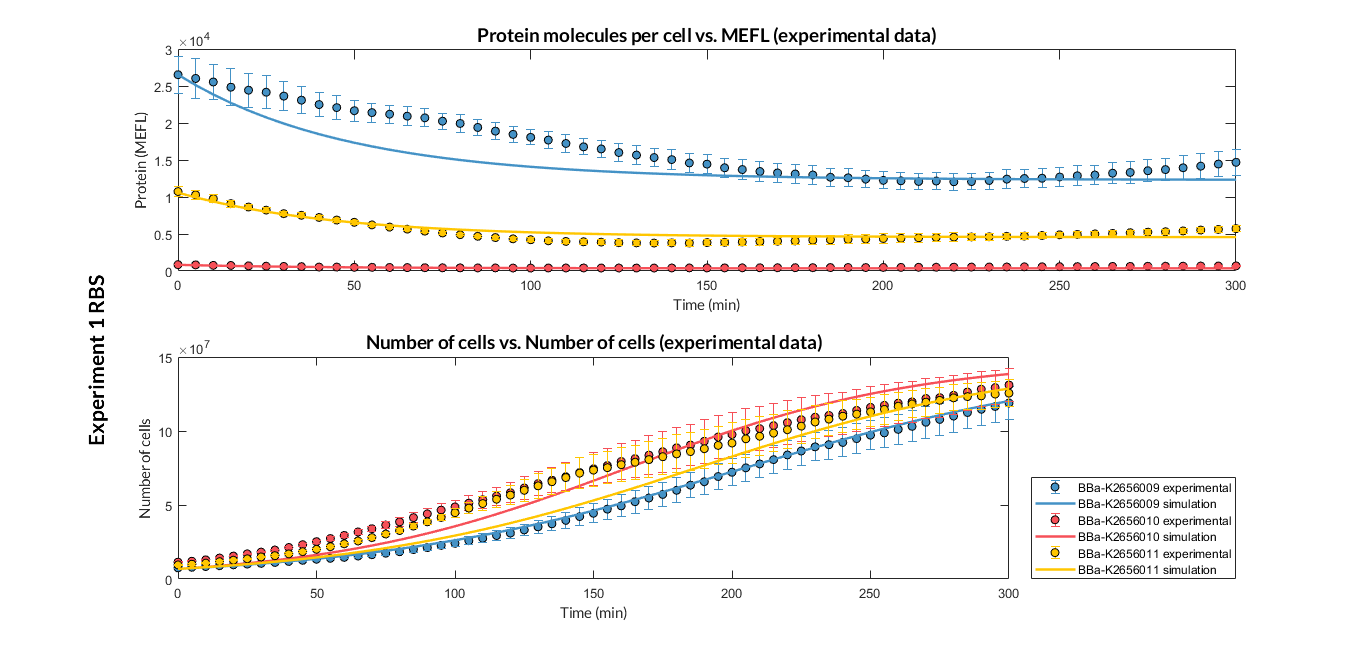
Characterization of the this part was performed with the transcriptional unit BBa_K2656101 , which was used in a comparative RBS expression experiment with composite parts BBa_K2656101 and BBa_K2656102. They all were assembled in a Golden Braid alpha1 plasmid using the same promoter, CDS and terminator.
By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model]and rationale choose its [http://2018.igem.org/Team:Valencia_UPV/Modeling#optimization optimization values] based on each RBS tested.
| Table 1. Optimized parameters from experimental data (BBa_K2656009 RBS). | |||
| Parameter | Value | ||
| Translation rate p | p = 0.082 min-1 | ||
| Dilution rate μ | μ = 0.01631 min-1 | ||
We have also calculated the relative force between the different RBS, taking BBa_K2656009 strong RBS as a reference. Likewise, a ratio between p parameters of the different RBS parts and p parameter of the reference RBS has been calculated.
| Table 2. BBa_K2656008 (GB BBa_B0032 RBS) relative strength and p ratio. | |||
| Parameter | Value | ||
| Relative strength | 0.371 | ||
| p parameter ratio (pRBS/pref) | 0.398 | ||
Shanghai_HS_United 2019's characterization
BBa_B0034 RBS (Elowitz 1999) -- defines RBS efficiency
All the construction is type IIS standard assembly. The different scar may effect the RBS efficiency for expression.
culture
Material: LB medium (2mL), pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP, pSB1K3-GFP, pSB1K3-LacZ (BBa_K3136014, BBa_K3136015, BBa_K3136017, BBa_K3136007)
1. Take several test tubes and add to 2mL Kan medium respectively.
2. Take the cultured pSB1K3-GFP, pSB1K3-LacZ, pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP, use the needle tip to take a small amount, and put it onto the shaking table.
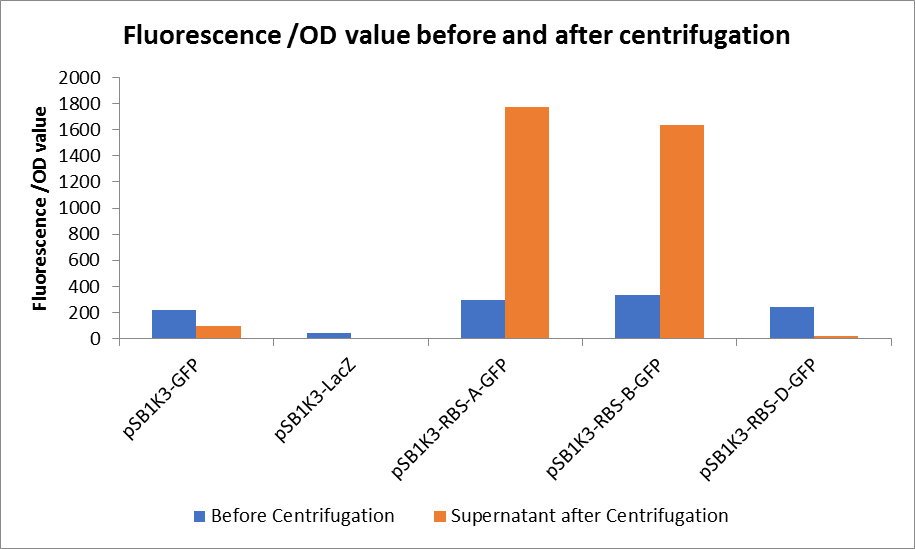
3. Measure the fluorescence value and the OD value before and after centrifugation.
Results:
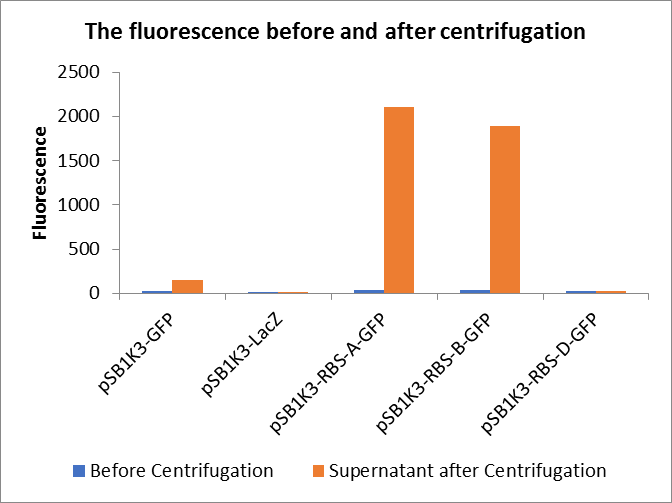
In this experiment, RBS (Elowitz 1999) -- defines RBS efficiency is same as pSB1K3-GFP.
Through series of procedures, we have measured the fluorescence value before and after centrifugation (Fig1). The fluorescence of pSB1K3-GFP, pSB1K3-LacZ and pSB1K3-GFP, pSB1K3-LacZ have no differences whatever before or after centrifugation. But after centrifugation, the fluorescence of pSB1K3-RBS-A-GFP and pSB1K3-RBS-B-GFP all increased substantially.
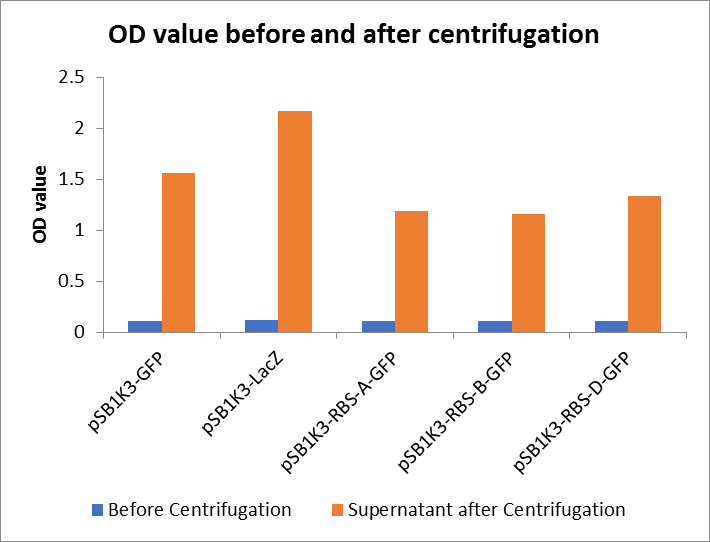
As shown in Fig2, the OD value of all samples before centrifugation was always lower than after centrifugation. Without the background interference of OD value, we can see the results clearly(Fig3). The fluorescence of pSB1K3-GFP hardly changed. It demonstrated that pSB1K3-GFP expressed a little. The intensity of RBS site in pSB1K3-GFP was weak.
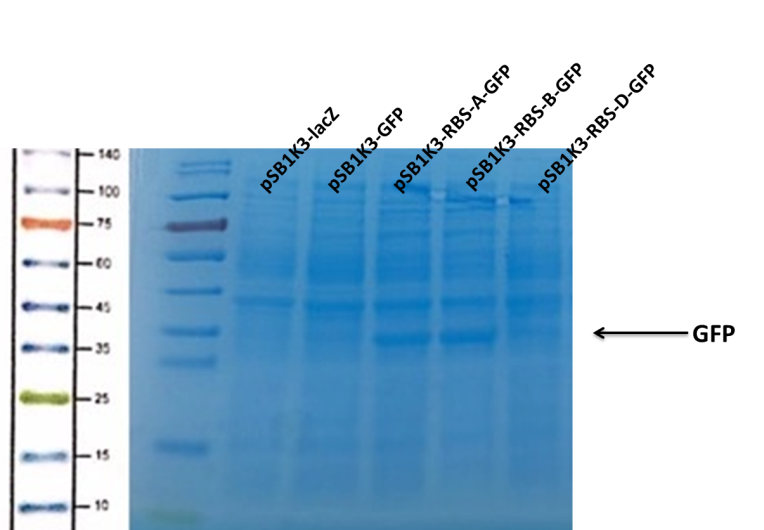
(1) Protein Electrophoresis
Material: pSB1K3-LacZ, pSB1K3-GFP, pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP, loading solution (6 μL), deionized water (1000 μL), marker solution, Coomassie brilliant blue dye. Note: pSB1K3-LacZ, pSB1K3-GFP, pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP are used for collaboration with the team Shanghai_HS_United, and these plasmids were expression in strain E. coli DH10B.
1. Add 24 μL pSB1K3-LacZ, pSB1K3-GFP, pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP, to 1.5mL tube respectively, and then add 6 μL loading.
2. Add 500 μL deionized water and centrifuge the tubes respectively.
3. Add another 500 μL deionized water, blow and suspend the solution.
4. Put the tubes 99℃ water for 15 minutes.
5. After heating, centrifuge the tubes for 5min.
6. Take and install the gel, add the electrophoresis buffer to the sample hole, and check if there is any leakage. Add marker, to the first and last sample holes, add pSB1K3-LacZ,pSB1K3-GFP and pSB1K3-RBS-A-GFP, pSB1K3-RBS-B-GFP, pSB1K3-RBS-D-GFP, MG1655 bpul, DH10B bpul, BL21(DE3) bpul to the other sample holes, respectively.
7. Connect the electrophoresis device to the power supply. Connect the positive electrode to the tank and the negative electrode to the slot for electrophoresis. The voltage is adjusted to 160V.
8. Turn off the power supply and disconnect the electrode until the bromophenol blue reaches the bottom of the release adhesive. Remove the glass sheet from the electrophoresis device and then remove the gel.
9. Soak the gel in Coomassie brilliant blue dye and dye it in a horizontal shaking bed for 15 min.
Observe the protein bands.
We can see that the lane of pSB1K3-GFP have no band(Fig4) at the location of GFP. It has been showed a low expression of pSB1K3-GFP and a feeble intensity of the RBS. Thus, the intensity of RBS was very important for the expression of proteins. The higher the intensity of RBS was, the more proteins expressed.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
Improvement from iGEM 2021 SZPT-China
Biology
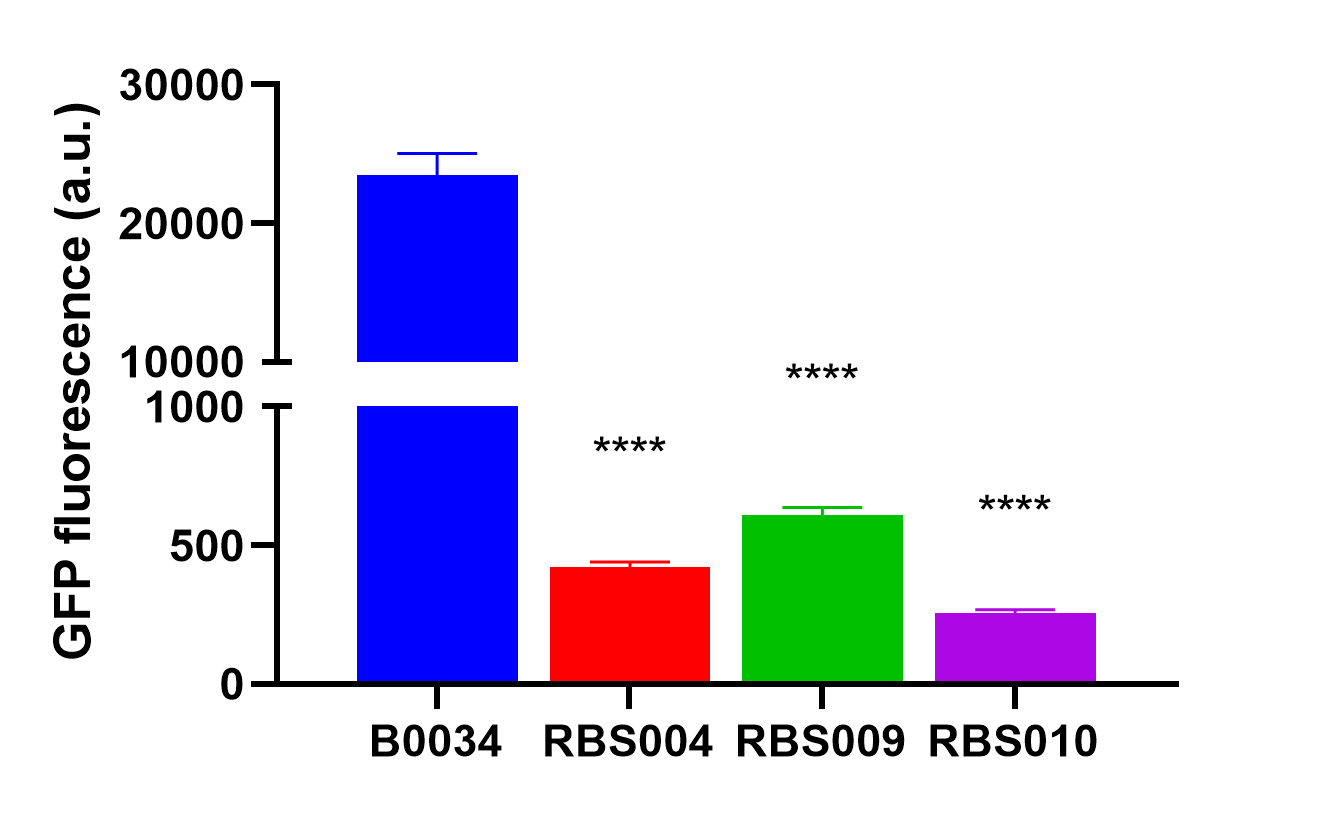
Fluorescence quantification was performed using the B0034 construct J23100-B0034-sfGFP-rrnB T1 (BBa_K3740058).
Usage
by the abovemetioned RBS004 (BBa_K3740018), RBS009 (BBa_K374008), and RBS010 (BBa_K3740010) to obtain an array of resultant plasmids as listed in Table 1. These plasmids were then transformed into E. coli DH5α, we quantified the fluorescence intensity when the optical absorbance of bacterial culture at 600nm was around 0.2. And were analyzed with the fluorescent proteins expressed by RBS004, RBS009, and RBS010, respectively.
Characterization
The average fluorescence intensity of sfGFP induced by B0034.
References
[1] Zhang H M , Chen S , Shi H , et al. Measurements of Gene Expression at Steady State Improve the Predictability of Part Assembly[J]. Acs Synthetic Biology, 2016, 5(3):269.
Improvement from iGEM Athens 2022
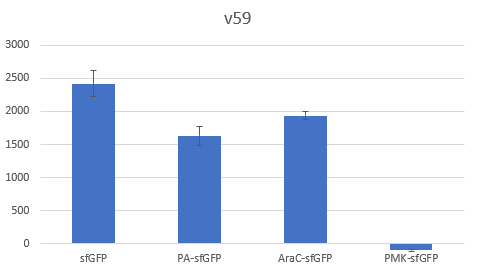
The context dependency of the BBa_B0034 RBS was improved by changing the biobrick spacer scar between the RBS and the ATG start codon, thus creating the RBS V59 BBa_K4294559. V59 was the result of an in silico analysis described thoroughly in our wiki page. Original BBa_B0034 as it is distributed by iGEM: tactagag aaagaggagaaa tactaa BBa_K4294559 : tactagag aaagaggagaaa ccctcca
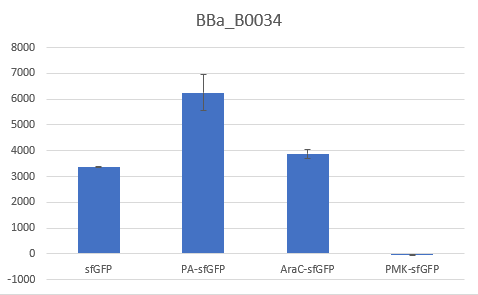
To verify the reduction in context dependency of the improved RBS spacer variants, different genetic contexts were introduced by fusions of the fluorescent protein sfGFP with the first 36 nucleotides of the proteins Penicillin acylase (PA) (BBa_K4294203), Phosphomevalonate Kinase (PMK) (BBa_K4294204) and AraC (BBa_K4294205).
The above parts together with the CDS of the sfGFP (BBa_K429420) create four genetic contexts in which the original BBa_B0034 RBS and the V59 RBS BBa_K4294559 variant were tested. Unfortunately, the 36ntPMK - sfGFP fusion did not produce any fluorescence in both groups and therefore the context dependency was evaluated in three genetic contexts. The genetic circuits were engineered to be controlled by the pTet promoter and tested in DH5a-z1 cells, which have genome-encoded TetR expression. 0.5 μM of aTc was used for induction.
Bar Plot of the Fluorescence/OD (in arbitrary units) ratios of the different reporters under the translational control of the BBa_B0034 RBS after a 3h induction with 0.5 μM of aTc. The 36nt PMK - sfGFP fusion was proven not to function in both RBS groups and was excluded from the CV calculation.
Bar Plot of the Fluorescence/OD (in arbitrary units) ratios of the different reporters under the translational control of the V59 BBa_K4294559 RBS after a 3h induction with 0.5 μM of aTc. The 36nt PMK - sfGFP fusion was proven not to function in both RBS groups and was excluded from the CV calculation.
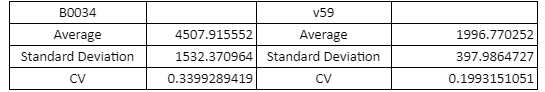
Table with Average, Standard Sample Deviation and CV values of the two RBS groups
The V59 RBS the Coefficient of Variance regarding the output signal is reduced by ~14%. The total output is also decreased; it is however in the same order of magnitude in comparison with the output of the BBa_B0034 regulated reporters.
The BBa_B0034 RBS was improved regarding its context dependence by the alteration of the RBS - Start Codon spacer region and the creation of the V59 BBa_K4294559. An obvious caveat regarding our results is the limited sample of the tested genetic contexts. However, since the choice of the potential spacer regions was supported by the in silico analysis, we believe that the V59 RBS is indeed an attractive alternative of the BBa_B0034 RBS and we encourage future iGEM teams in further characterizing it in new genetic contexts alongside with the other RBS candidate sequences that were not tested by our team. The potential reported lower translation rate could be alleviated by transcriptional enhancers leading to the reconstruction of existing RBS parts that will achieve similar average translation rates in comparison with their respective original parts, but with a lower context dependence.
Combine BBa-B0034 with PPK1 Nanjing-China 2024
iGEM Nanjing China 2024
We used the part for our reporter system and it worked very good!It can be used as a good RBS in S.oneidensis, which we have used extensively in our projects.
We developed RBS of different intensities in tandem with PPK1 and expressed them in S.oneidensis.This RBS element works well in S.oneidensis.

Figure 1: Basic construction of RBS-PPK1 plasmid
The best results can be seen after adopting the RBS BBa-B0034, which is a function-based verification.

Figure 2: Different phosphorus accumulation capacity in S. oneidensis. using different RBS
We also performed polyacrylamide gel electrophoresis and found that there was indeed the largest amount of PPK1 expression in tandem with the B0034 element, which was a direct validation based on protein expression.

Figure 3: Different levels of protein expression in S. oneidensis. using different RBS
B0030 vs B0034 in E.coli BL21 (DE3) - Thessaly 2024
Group: Thessaly 2024
Purpose
BBa_B0030 and BBa_B0034 were the RBSes that we used in our promoter testing experiments. We wanted to check which RBS would be the strongest and which would be the ideal candidate for our design.
Methods
Measurements for OD 600nm and fluorescence (488nm,515nm) were taken over the course of 16 hours, in a 96-well microplate. Clonings were done according to the Golden Braid method, leaving us with level α constructs in the pDGB3a1 backbone BBa_K4213058.
Constructs and pDGB3a1 (empty vector) transformed into E.coli BL21 (DE3) chassis, incubated at 37oC, 180rpm for 16 hours.
Here you can find more information about the constructs tested containing BBa_B0030: BBa_K5299200, BBa_K5299202,BBa_K5299204.
Here you can find more information about the constructs tested containing BBa_B0034: BBa_K5299201, BBa_K5299203,BBa_K5299205.
Plated 200 ul 5 times, out of each single liquid bacterial culture, created from the same bacterial colony, in order to establish accuracy through technical repeats.
Medium used was M9 due to minimal interference, with D-glucose serving as the carbon source. Also, served as blank, plated 5 times.
Measurements were normalised as such: using the average price of fluorescence for the 5 technical repeats and dividing it by the average price of OD.
Standard deviation included in the graphs.
Results
Measurements were taken over the course of 16 hours.
Here are the results for BBa_B0034 regarding BBa_J23119 in the pDGB3a1 backbone. BBa_B0034 proved stronger than BBa_B0030.

For more information regarding BBa_J23119, head over to BBa_J23119.
Here are the results for BBa_B0034 regarding BBa_J45992 in the pDGB3a1 backbone. BBa_B0034 proved stronger than BBa_B0030.

For more information regarding BBa_J45992, head over to BBa_J45992.
Here are the results for BBa_B0034 regarding P3.1. (BBa_K4583008) in the pDGB3a1 backbone. BBa_B0034 proved stronger than BBa_B0034.

For more information regarding P3.1, head over to BBa_K4583008.
Multiple B0034 in a Composite Part - SHSBNU-China 2024
Group: SHSBNU-China 2024
In 2024, our team SHSBNU-China built a composite part including 5 functional enzymes, BsAld, CsAlaDC, GMAS, PPK, and GNP1 to facilitate the de novo synthesis of L-theanine in E. coli, using simple glucose as the input substrate. </br> In order to have homogeneous expression of all the functional enzymes, all the coding gene were arranged in a series, all in between a T7 promoter and the terminator. Each coding sequence was led by its own RBS, part number BBa_B0034. </br> The results provided evidence that in a composite vector including multiple parts, using the B0034 RBS before each part can work as expected to produce functional enzymes.


References
[1] Hagihara R, Ohno S, Hayashi M, Tabata K, Endo H. Production of l-Theanine by Escherichia coli in the Absence of Supplemental Ethylamine. Appl Environ Microbiol. 2021 May 11;87(11):e00031-21. doi: 10.1128/AEM.00031-21.
[2] Cao, R., Hu, S., Lu, Y., Wang, W., Fu, Z., & Cheng, J. (2023). Fermentative Production of L-Theanine in Escherichia coli via the Construction of an Adenosine Triphosphate Regeneration System. Fermentation, 9(10), 875–875.
//direction/forward
//function/coliroid
//rbs/prokaryote/constitutive/community
//regulation/constitutive
//ribosome/prokaryote/ecoli
| biology | |
| efficiency | 1 |

 1 Registry Star
1 Registry Star