Part:BBa_K2273023
AmyE signal peptide of B. subtilis alpha-amylase
The AmyE signal peptide is part of the Signal Peptide Toolbox of [http://2017.igem.org/Team:TU_Dresden iGEM Team TU Dresden 2017 (EncaBcillus - It's a trap!)].
The signal peptide (amino acids 1 to 33) of Bacillus subtilis alpha-amylase (starch degradation, Uniprot [http://www.uniprot.org/uniprot/P00691 P00691]) targets for protein secretion via the Sec-SRP secretion pathway (Brockmeier et al, 2006).
This part was generated in a modified version of RFC25, where a strong Shine Dalgarno Sequence (SD) is included, and has the following prefix and suffix:
| Prefix with | EcoRI, NotI, XbaI and SD | 5'-GAATTCGCGGCCGCTTCTAGATAAGGAGGTCAAAA-3' |
| Suffix with | AgeI, SpeI, NotI and PstI | 5'-ACCGGTTAATACTAGTAGCGGCCGCTGCAGA-3' |
Sites of restriction enzymes generating compatible overhangs are indicated by sharing one color. (EcoRI and PstI are marked in blue, NotI in green, XbaI and SpeI in red and AgeI in orange. Additionally, the Shine-Dalgarno sequence is marked in silver and the stop codon is underlined.)
This part is used in the 2017 TU Dresden iGEM project [http://2017.igem.org/Team:TU_Dresden EncaBcillus - It's a trap!] and part of the Signal Peptide Toolbox.
Signal Peptide Toolbox
The GRAM-positive model organism B. subtilis is considered a perfect host for heterologous and recombinant protein secretion due to its extracellular chaperones, natural protein secretion capacity and well-studied genetics (van Dijl and Hecker, 2013). To increase protein production rates of such proteins, it is feasible to enhance the secretion efficiency. The easiest method to realise this, is to tag the protein of interest with a so-called signal peptide of B. subtilis' general protein secretion pathway Sec-SRP which dictates the secretion of proteins into the surrounding supernatant of the cell (Fu et al, 2007).
Though the Sec-SRP protein secretion pathway of B. subtilis contains more than 170 distinct signal peptides, every single signal peptide varies in its secretion efficiency in dependency on the protein fused to it. To date, it is impossible to predict this dependency via studying the genetic code of the signal peptides and the protein of interest (Brockmeier et al, 2006).
To cope with this issue, the [http://2017.igem.org/Team:TU_Dresden iGEM team of TU Dresden 2017 (EncaBcillus - It's a trap!)] developed a novel shotgun approach high-throughput signal peptide screening vector system for targeted secretion in B. subtilis, or âSignal Peptide Toolboxâ for short.
The Signal Peptide Toolbox contains the so-called Evaluation Vector and the Signal Peptide Mix which is made of equal amounts of all signal peptides which are part of the Signal Peptide Toolbox. It can be used by any team to increase secretion efficiency of heterologous and recombinant proteins in B. subtilis.
The iGEM team TU Dresden 2017 provides an Evaluation Vector construct with an inducible promotor. Nonetheless, the vectors' promotor can still be replaced easily.
For screening, in a first step, the coding sequence of the protein of interest as well as a promotor of choice would be cloned into the Evaluation Vector. In a second step, all signal peptides of the Signal Peptide Mix would be integrated into the vector at once. Finally, the randomly transformed bacteria colonies are screened for their specific secretion efficiency using a protein of interest specific assay. Those strains, which reassemble the secretion level of choice the best, can then be sequenced to reversely identify the integrated signal peptide. This process is shown in the following video.
The following list of signal peptides gives an overview of all signal peptides which are currently part of the Signal Peptide Mix of the Signal Peptide Toolbox.
Sequence and Features
The DNA sequence of this part has been verified via sequencing before it was sent in.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
Our team, Technion 2019 (âBeeFreeâ), used this part to secrete the Glucose Oxidase (GOx) enzyme from Bacillus subtilis.
Our composite part, BBa_K2934004, composes of the AmyE secretory signal peptide, a short peptide linker (BBa_K2934005) and the enzyme GOx (BBa_K2934000).
We tested this composite part at our lab and showed a successful secretion and extracellular activity by preforming assays for both the supernatant and the lysate of our engineered bacteria (for further information, see methods below).
Glucose Oxidase Enzymatic Activity Assay
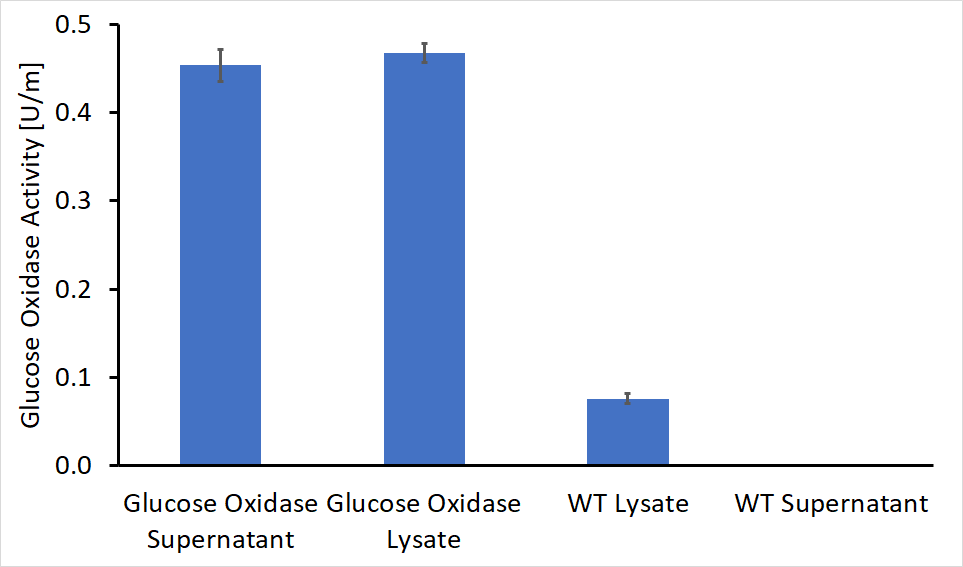
After separating the bacteria supernatant and lysate, we concentrated the proteins in both solutions using a HisTag column. Following concentration, we performed enzymatic activity assays for both the lysate and the supernatant, compared to the wild type bacteria. The results of these experiments are shown in Figure 1. As can be seen, GOx was active both inside the bacteria cell and as a secreted enzyme outside the bacteria. The results also show that roughly 50% of the produced protein was secreted from the bacteria to its surroundings.
As shown in figure 1, some GOx activity was detected in the wild type bacteria lysate, although this activity was significantly lower compared to the engineered bacteria. A possible explanation for this observation is the presence of other enzymes within the bacteria with similar activity or imprecise lab work.
Enzyme Secretion Assay Using SDS-PAGE
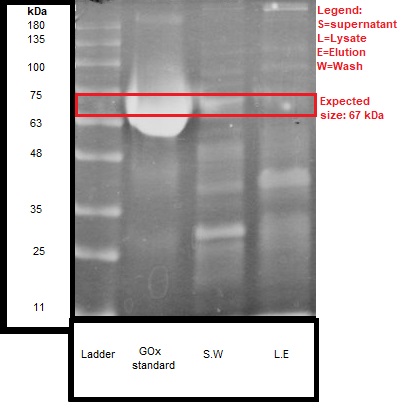
We ran the discussed samples on SDS-PAGE, and the results are shown in Figure 2.
We can see that the expected size of the protein (~67 kDa) was detected both in the lysate and the supernatant of the engineered bacteria.
These results give further confirmation, together with the activity test, that secretion of about 50% GOx enzyme was received. It should be noted that there were no detectable bands in the wild type bacteria samples.
Methods
This vector was constructed by Gibson assembly, with the pBE-S commercial plasmid (Takara) that served as a backbone, and eventually transformed into Bacillus subtilis (RIK1285).
The bacteria were cultured over-night at 37°C in a shaking incubator (250 rpm), followed by 1:100 dilution. The diluted culture was incubated for another few hours while monitoring the culture turbidity by measuring the O.D600.
When the culture has reached the logarithmic growth phase, the bacterial extracellular and intracellular proteins were separated, after collecting the supernatant and the pellet (followed by lysis, using sonicator).
//chassis/prokaryote/bsubtilis
| None |