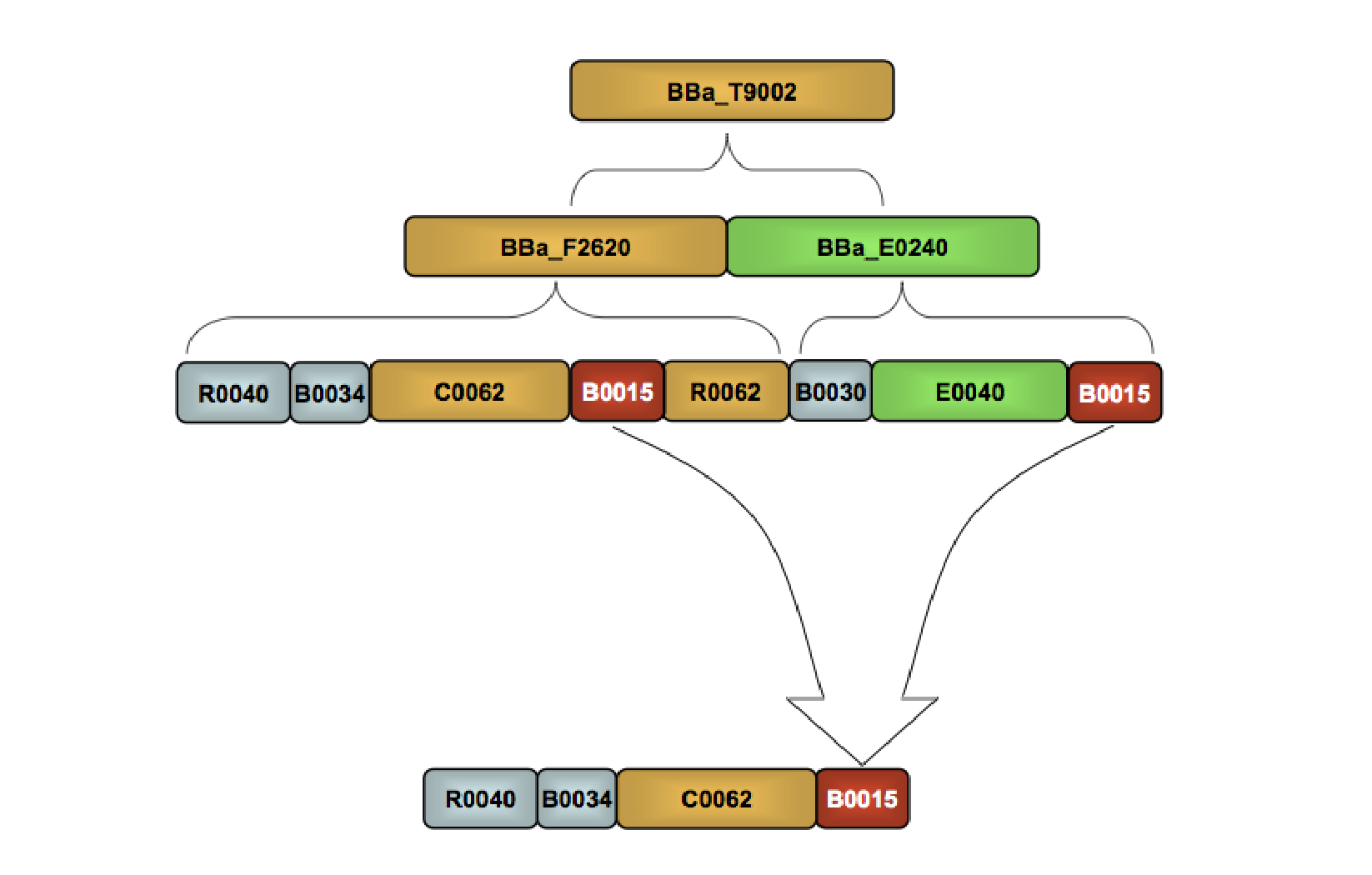
Part:BBa_F2620:Stability
|
|
Description
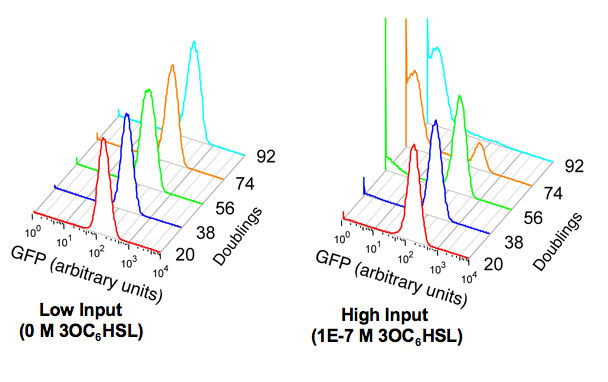
Device stability describes how the transfer function of the device changes across multiple rounds of cell division and culture. As with our other experiments, we used the fluorescent reporter device, BBa_E0240 to indirectly measure the output from BBa_F2620. We measured the performance of the device under both high and low input conditions over 94 doublings. Please see the part experience page for a general description of our characterization method.
Data
Protocols
- A 5 ml culture of [http://openwetware.org/wiki/Endy:M9_media/supplemented supplemented M9 medium] and antibiotic (kanamycin, 20 µg/ml) was inoculated with a single colony of BBa_T9002 from a fresh plate.
- The culture was grown in a 17 mm test tube for 15 hrs at 37°C with shaking at 70 rpm.
- The culture was diluted 1:400 into 5 ml of fresh medium and grown under identical conditions for a further 10 hrs. The culture was diluted 1:4096 into two identical 5 ml cultures. 3OC6HSL was added to one of the cultures to an input level of 1E-7 M.
- These two cultures were propagated in a similar manner with 1:400 and 1:4096 dilutions in the morning and evening respectively every day for 5 days.
- Each day, following the overnight incubation of cultures, a sample of the overnight cultures was stored in a 20% glycerol solution at –80°C to allow later sequencing. Samples to be sequenced were streaked on LB plates containing kanamycin (20 µg/ml). Plasmid DNA from five colonies on each plate was purified using a Qiagen Spin Miniprep Kit ([http://www1.qiagen.com/ Qiagen]). DNA was sequenced and analyzed as described earlier using standard primers internal and external to the receiver and reporter device (BBa_G00100, BBa_G00101, and G00602).
- Each morning, a second culture was inoculated by a 1:400 dilution from the overnight culture for both the high and low input condition. These second cultures were grown in the absence of 3OC6HSL for 8 hrs. This growth period diluted-out the accumulated GFP from the culture propagated in the presence of 3OC6HSL before assaying performance.
- Samples from both of the second copies were induced with a high input level of 3OC6HSL (1E-7 M) at 37°C with shaking at 70 rpm for 45 min. Single-cell fluorescence measurements were carried out on a FACScan flow cytometer ([http://www.bdbiosciences.com/home/ Becton-Dickinson]) with a 488 nm Argon excitation laser and 525 nm emission filter. During each flow-cytometer measurement, data was collected from 50,000 cells. 2 µl of Sphero fluorescent beads (0.87 µm ø, [http://www.spherotech.com/ Spherotech]) in 500 µl H2O was used as a control for experiment-to-experiment variation of cytometer performance. FACScan data were analyzed using Cell Quest ([http://www.bdbiosciences.com/home/ Becton-Dickinson]) and FlowJo ([http://www.flowjo.com/ FlowJo]).
- Genetic stability is defined as the number of replication events until a mutant becomes fixed in the population. The performance stability of a device is defined as the number of replication events until the majority of the devices in the population have lost the ability to respond correctly to an input. Both the genetic and the performance stability were estimated from the FACS data shown in Figure 1 and genetic stability was confirmed by sequence analysis.
Origin of stability mutant
Repeated experiments following the stability protocol (above) consistently resulted in the accumulation of the same mutant after a similar number of doublings. Sequence analysis of individual clones from the culture at the end of the stability experiment showed that the mutant had a deletion in BBa_T9002 between two 143 bp homologous regions (Figure 2). These results suggested that there was pre-existing genetic variation within each clone at the start of each stability experiment. We measured the growth rate of the complete device and the mutant when grown under the same conditions as in the stability experiment. Exponential doubling times of 59 and 54 min were calculated for BBa_T9002 and the mutant device respectively under high input conditions. A simple model (not shown) suggests that this doubling time difference is consistent with the observed population takeover by a pre-existing mutant within 74 doublings.
To test the hypothesis that pre-existing genetic variation is the source of the mutant device plasmid DNA was purified from four colonies grown on a plate streaked from the master glycerol stock of MG1655 bearing BBa_T9002 using a Qiaprep spin miniprep kit ([http://www1.qiagen.com/ Qiagen]). The purified DNA was used as template for PCR reactions. Several PCR products were detected by gel electrophoresis, with identical results for each template DNA sample. A band running at the speed expected for the mutant device was used as template for a further PCR reaction. The PCR product was sequenced and found to match the sequence of the mutant device.
To ensure the previous result was not due to an artifact of the PCR process, a further test for the presence of the mutant plasmid in the glycerol stock was performed. To enrich for the mutant plasmid, purified DNA from the glycerol stock was digested with restriction enzymes BsrG1 and HincII that recognize nucleotide sequences in BBa_T9002 but not the mutant (NEB). The digested DNA was purified using a QiaQuick PCR purification kit ([http://www1.qiagen.com/ Qiagen]) and used to transform chemically competent E. coli TOP10 cells ([http://invitrogen.com Invitrogen]). The transformed cells were plated on LB plates containing kanamycin (20 µg/ml) and 1E-7 M 3OC6HSL. A fraction of the colonies were green as expected for cells transformed with the full length device and the remainder of the colonies were not visibly green. Colony PCR reactions were performed on colonies that were not visibly green. The PCR products were analyzed by gel electrophoresis. The PCR product of all non-green colonies ran at the speed expected for the mutant device, while the PCR products of all green colonies ran at the speed expect for BBa_T9002. This result, together with the result of the PCR approach described above made it near certain that the mutant device was present on a minority plasmid population within individual cells in the glycerol stock. Consequently, the stability data quoted for BBa_T9002 are lower bounds, set by the time taken for the mutant plasmid to become fixed in individual cells and then in the population.
We then investigated whether occurrence of the mutant plasmid when transforming cells with BBa_T9002 was repeatable. We purified DNA from visibly green colonies used in the experiment described above. The parent cell of each colony should be transformed with a single plasmid based on the transformation protocol used and hence the colony should contain a homogenous plasmid population. The purified DNA was analyzed similarly to that described above and the results again showed that the transformed cells carried a small fraction of mutant plasmids. This data suggests that the mutant occurs at a high frequency in both recA+ (MG1655) and recA mutant cells (TOP10), consistent with a replication slippage mechanism for repeat deletion.

 1 Registry Star
1 Registry Star