Difference between revisions of "Part:BBa I14017:Experience"
(→Background information) |
(→Modeling crosstalk) |
||
| Line 31: | Line 31: | ||
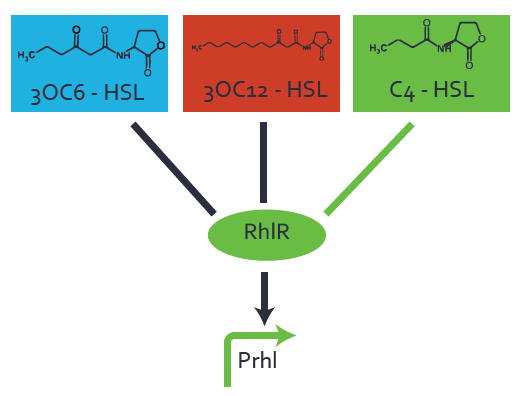
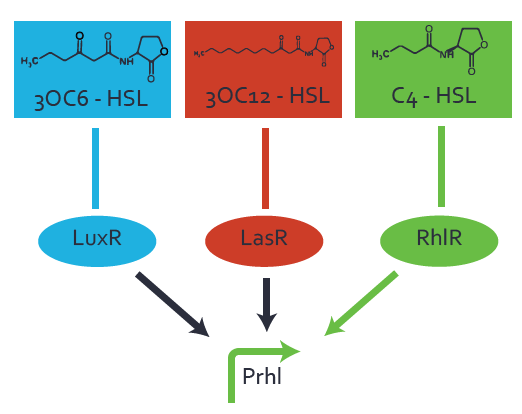
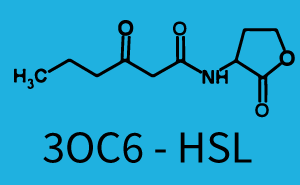
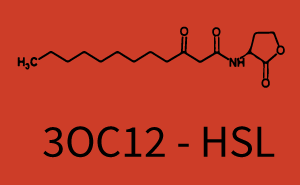
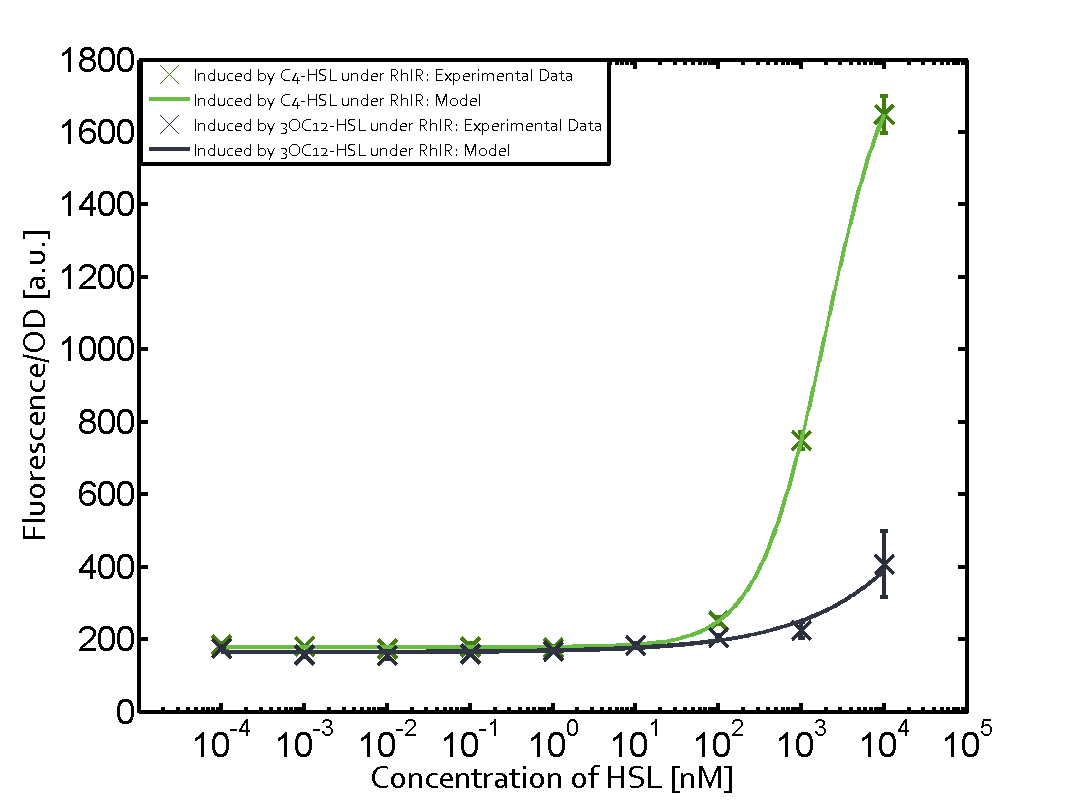
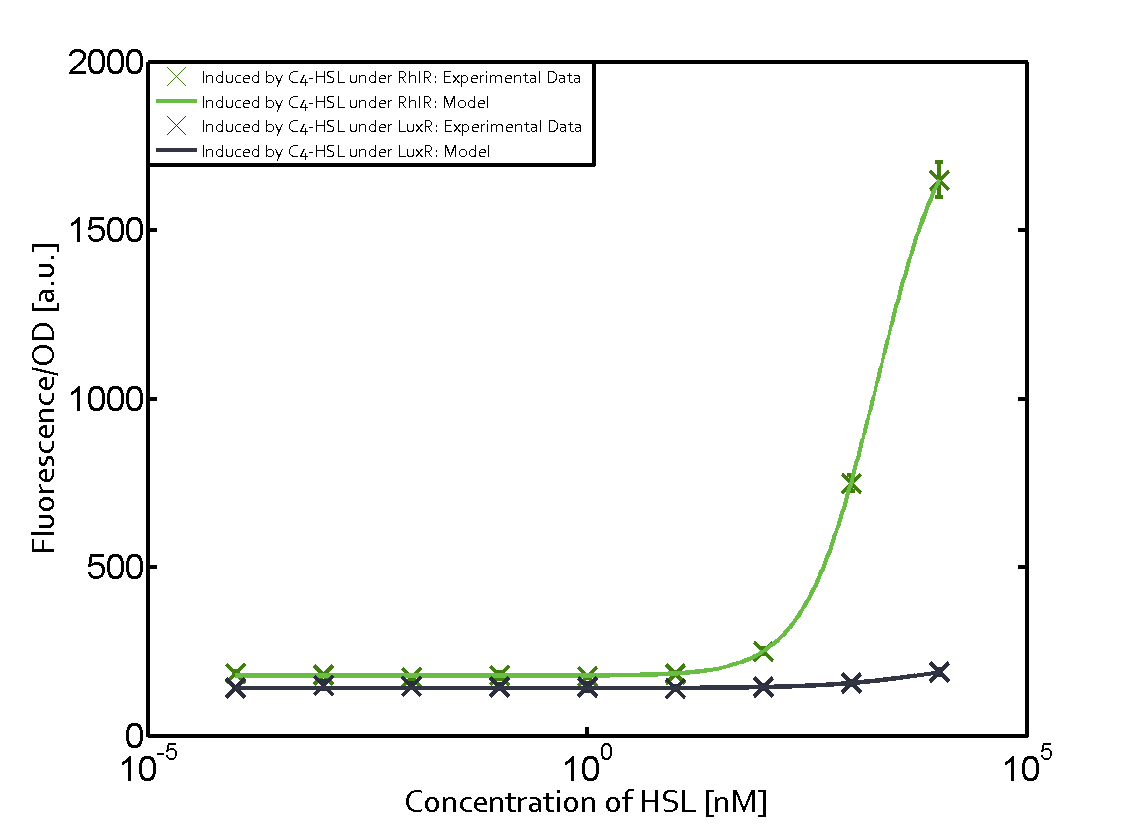
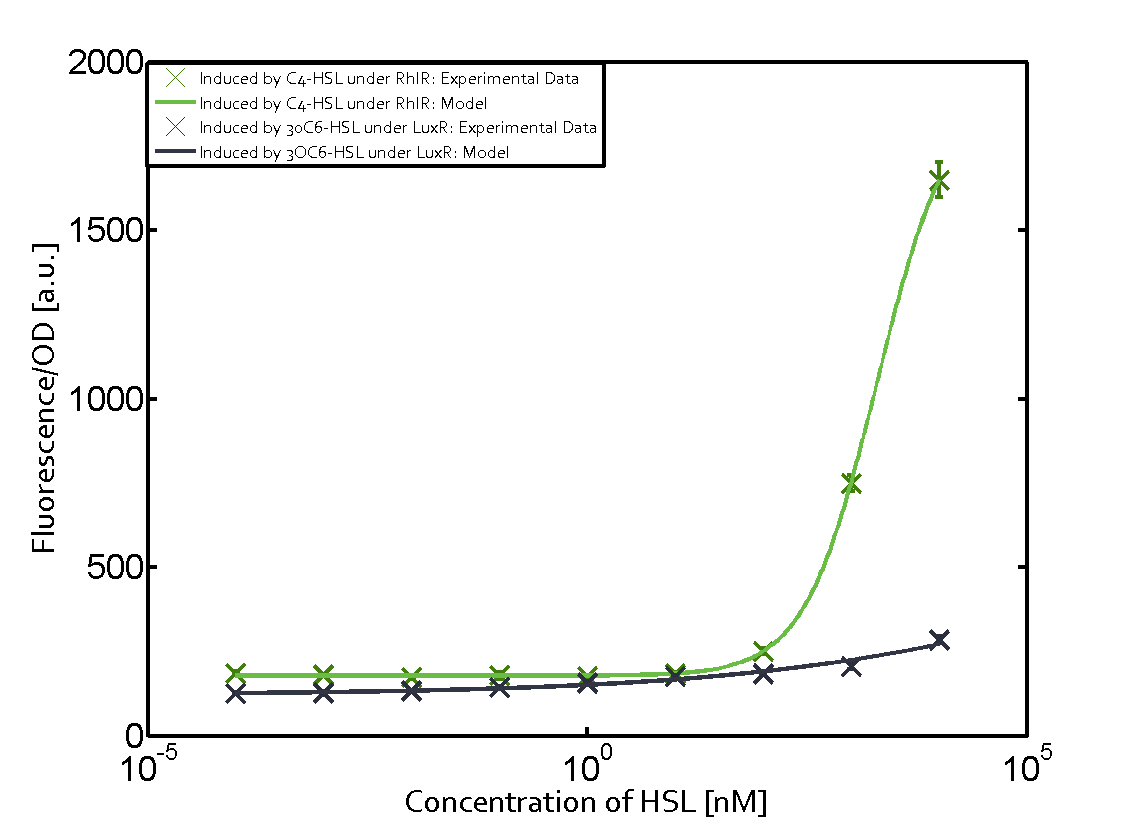
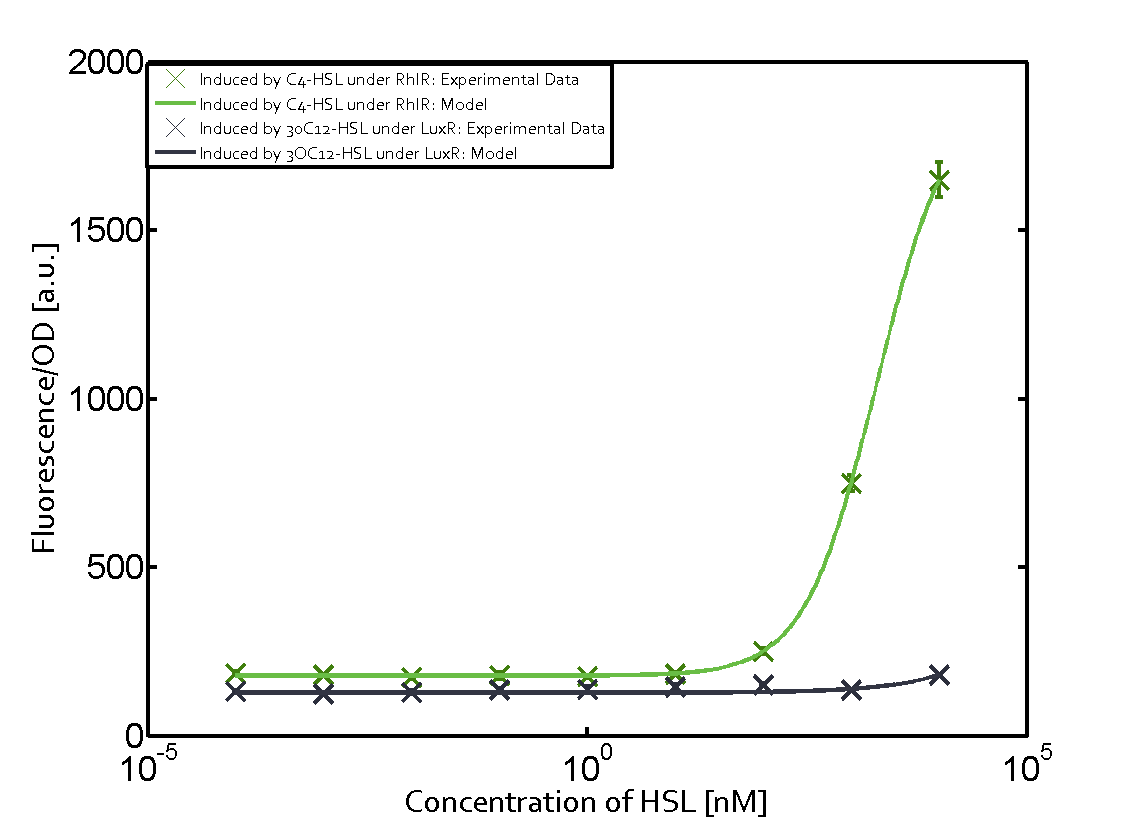
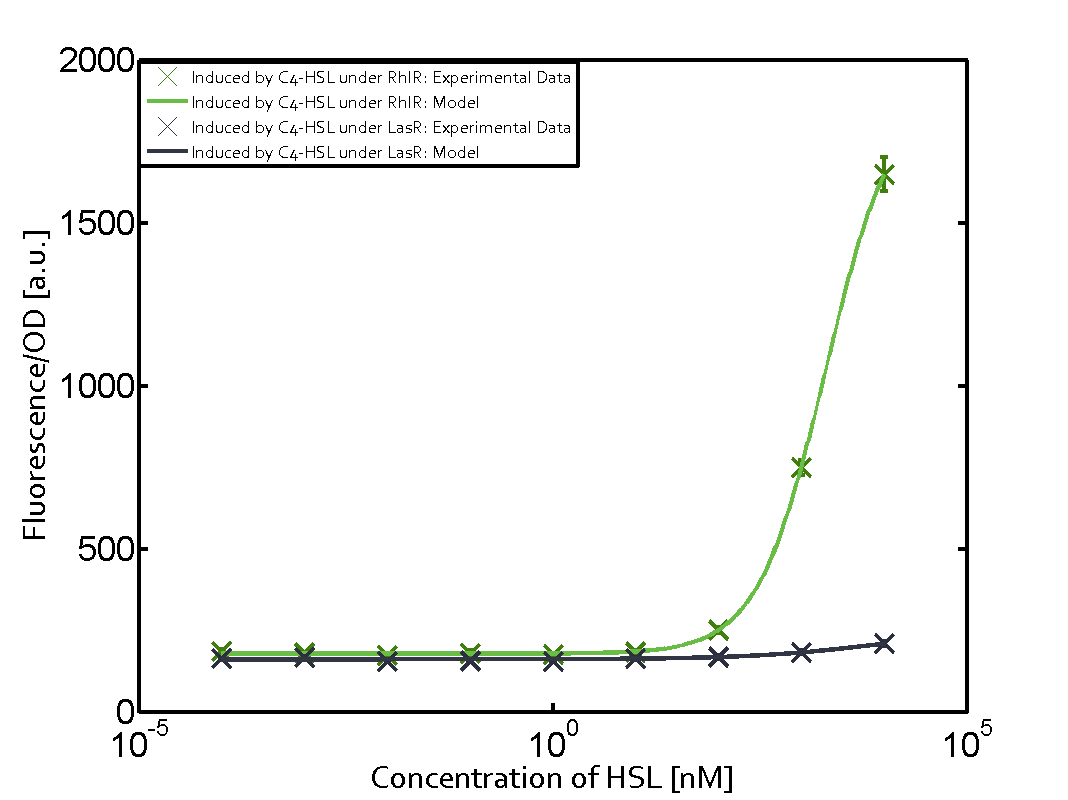
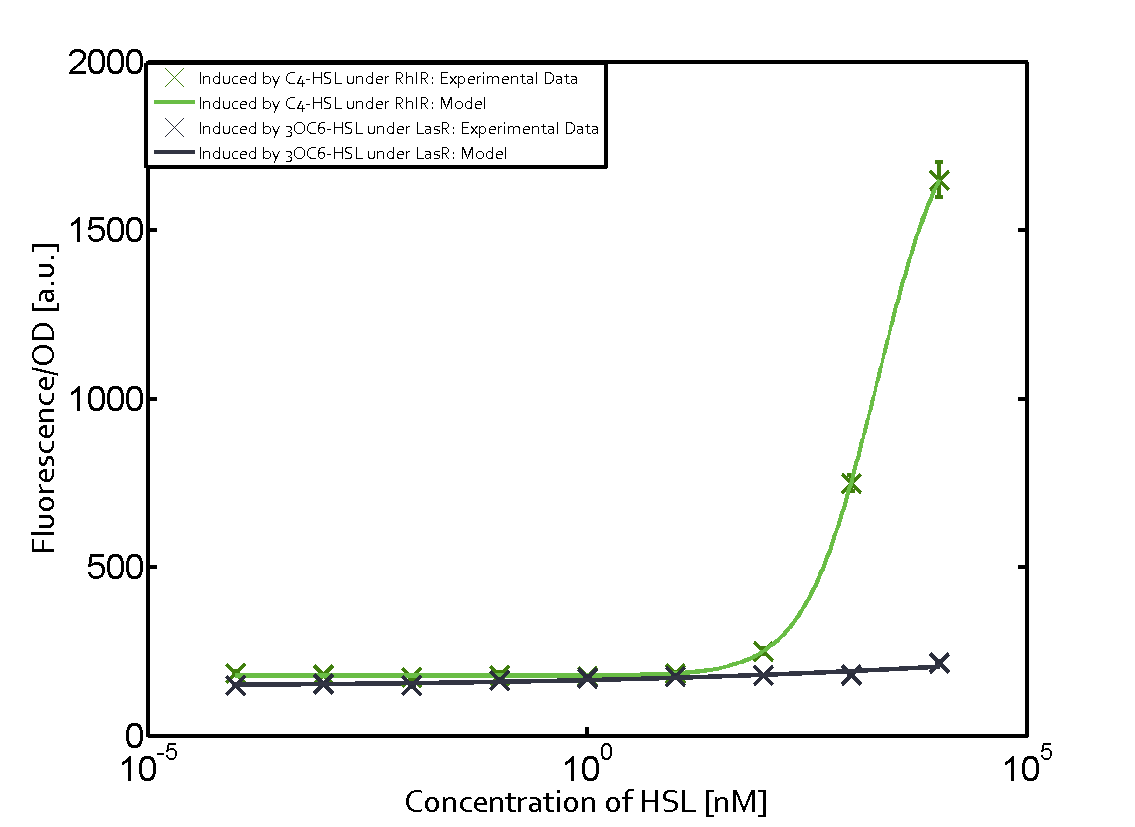
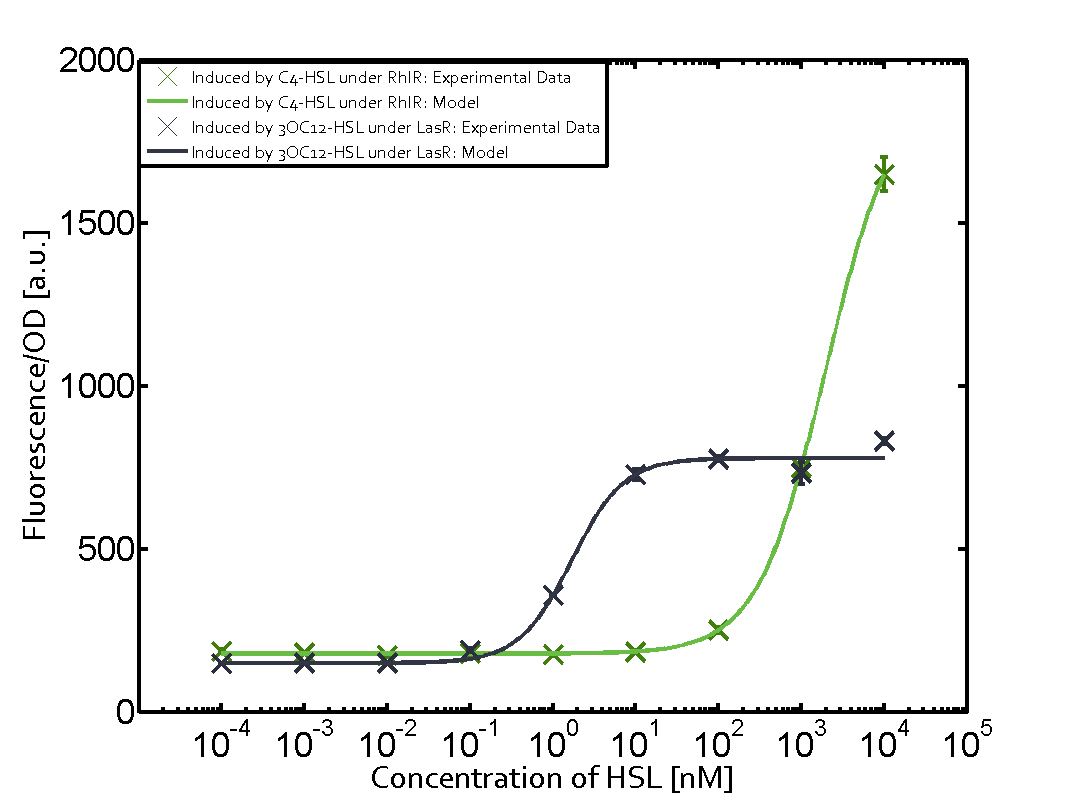
Ideally, C4-HSL (inducer) binds to RhlR (receptor) and this complex then activates the transcription of the gene regulated by Prhl. However, when using different inducers (3OC6-HSL and 3OC12-HSL) and receptors (LuxR and LasR), they can cross react to form complexes which activate the transcription of the gene regulated by Prhl. The effect of these possible combinations on the promoter have been characterized in detail below. | Ideally, C4-HSL (inducer) binds to RhlR (receptor) and this complex then activates the transcription of the gene regulated by Prhl. However, when using different inducers (3OC6-HSL and 3OC12-HSL) and receptors (LuxR and LasR), they can cross react to form complexes which activate the transcription of the gene regulated by Prhl. The effect of these possible combinations on the promoter have been characterized in detail below. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
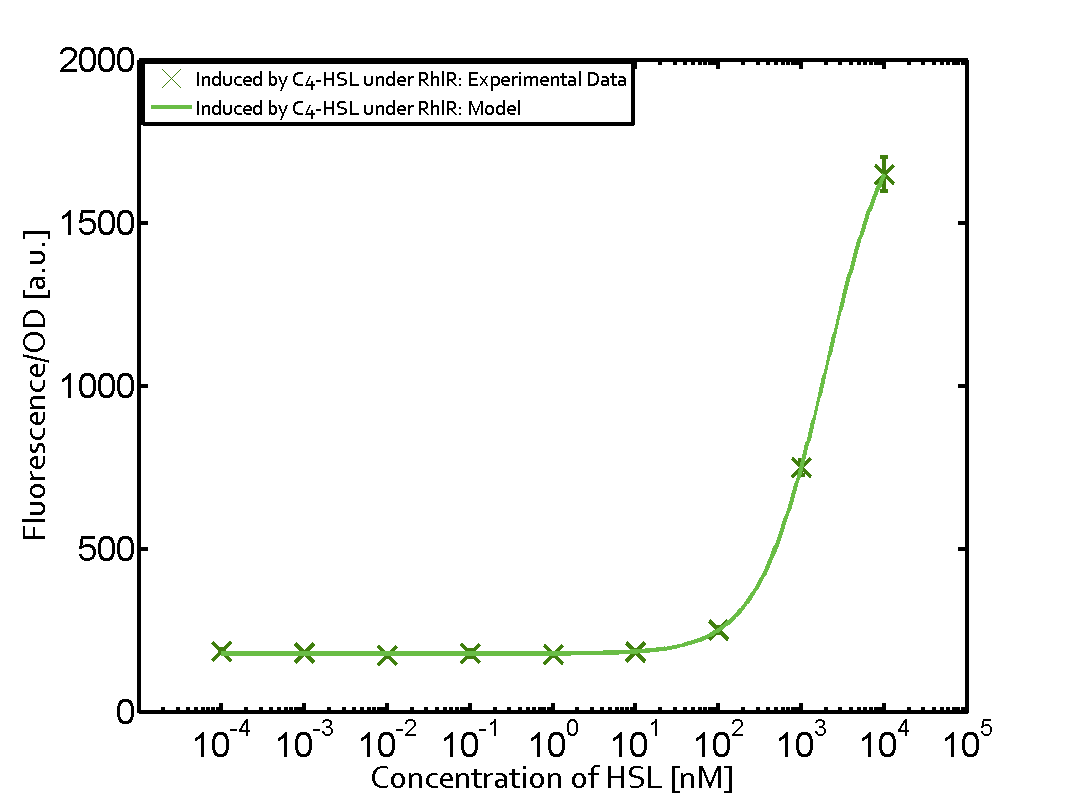
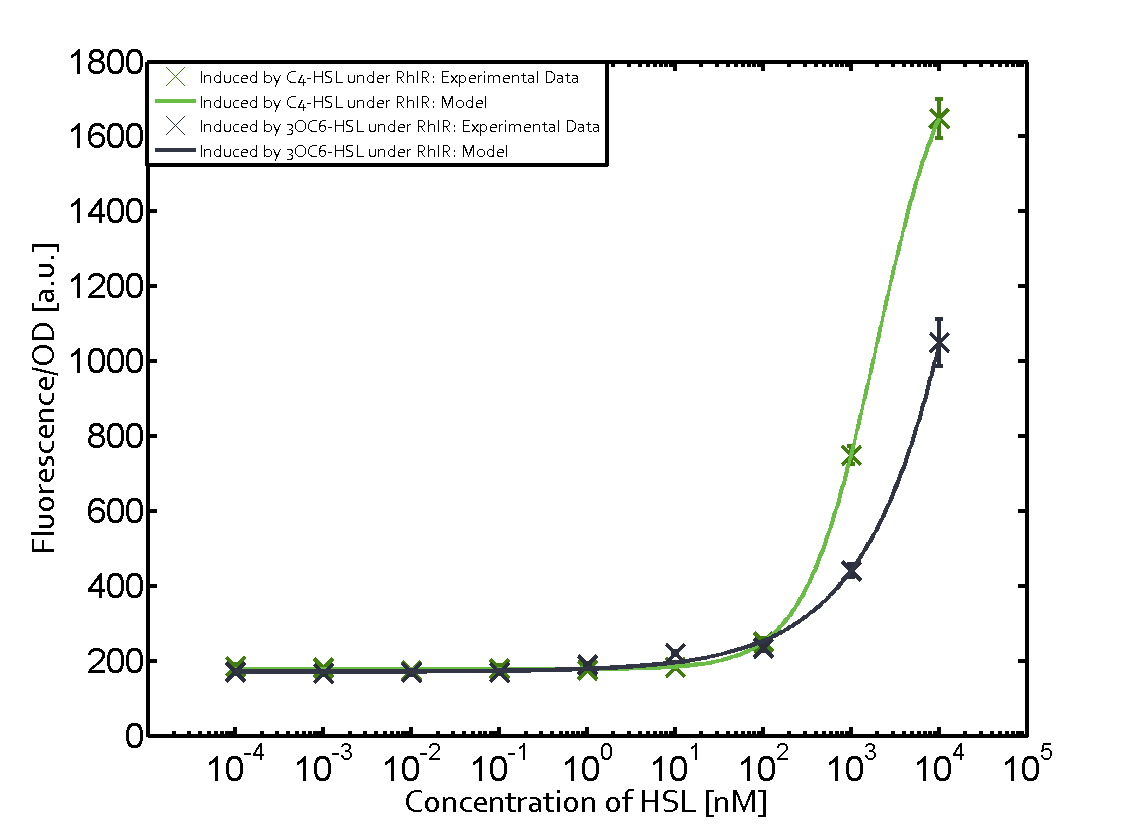
== First-order crosstalk == | == First-order crosstalk == | ||
Revision as of 13:43, 24 October 2014
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_I14017
User Reviews
UNIQ5e88bceeae1b5f79-partinfo-00000000-QINU
|
••••
ETH Zurich 2014 |
Characterization of two-order crosstalk on the promoterBackground informationIdeally, C4-HSL (inducer) binds to RhlR (receptor) and this complex then activates the transcription of the gene regulated by Prhl. However, when using different inducers (3OC6-HSL and 3OC12-HSL) and receptors (LuxR and LasR), they can cross react to form complexes which activate the transcription of the gene regulated by Prhl. The effect of these possible combinations on the promoter have been characterized in detail below. First-order crosstalkFirst Level crosstalk: RhlR binds to different HSL and activates the promoterSecond Level crosstalk: other regulatory proteins, like LuxR, bind to their natural HSL substrate and activates the promoterSecond order crosstalk: Combination of both cross-talk levelsOther regulatory proteins, like LuxR, bind to different HSL and activates the promoter. Results
| ||||||||||||||||||||
|
Antiquity |
This review comes from the old result system and indicates that this part did not work in some test. |
UNIQ5e88bceeae1b5f79-partinfo-00000003-QINU