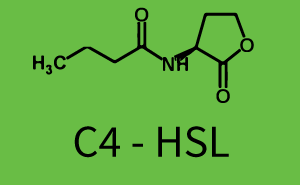
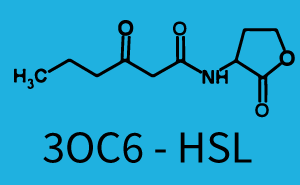
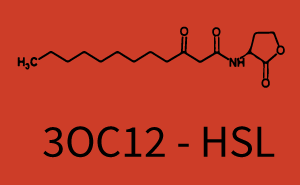
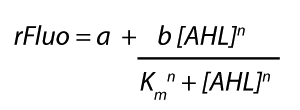Difference between revisions of "Part:BBa C0171:Experience"
(→Modeling crosstalk) |
(→Modeling crosstalk) |
||
| Line 37: | Line 37: | ||
[[File:ETHZ_HillEq.png|center|200px]] | [[File:ETHZ_HillEq.png|center|200px]] | ||
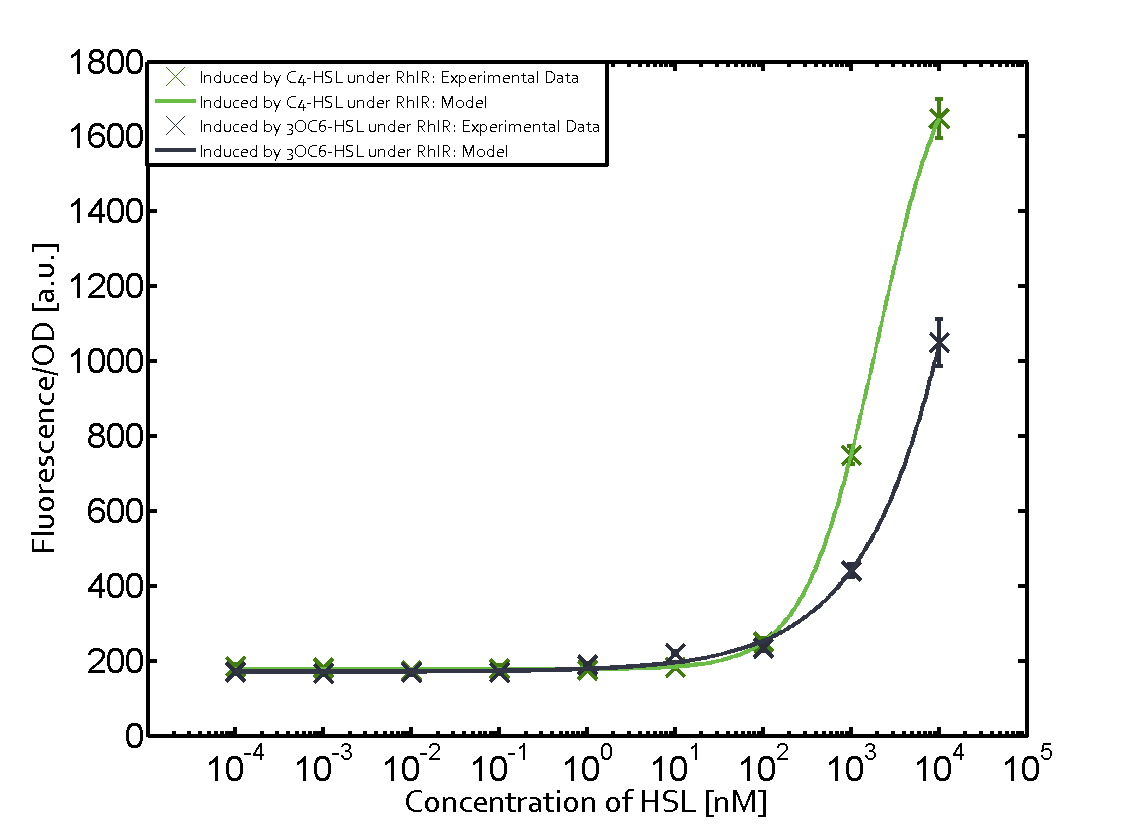
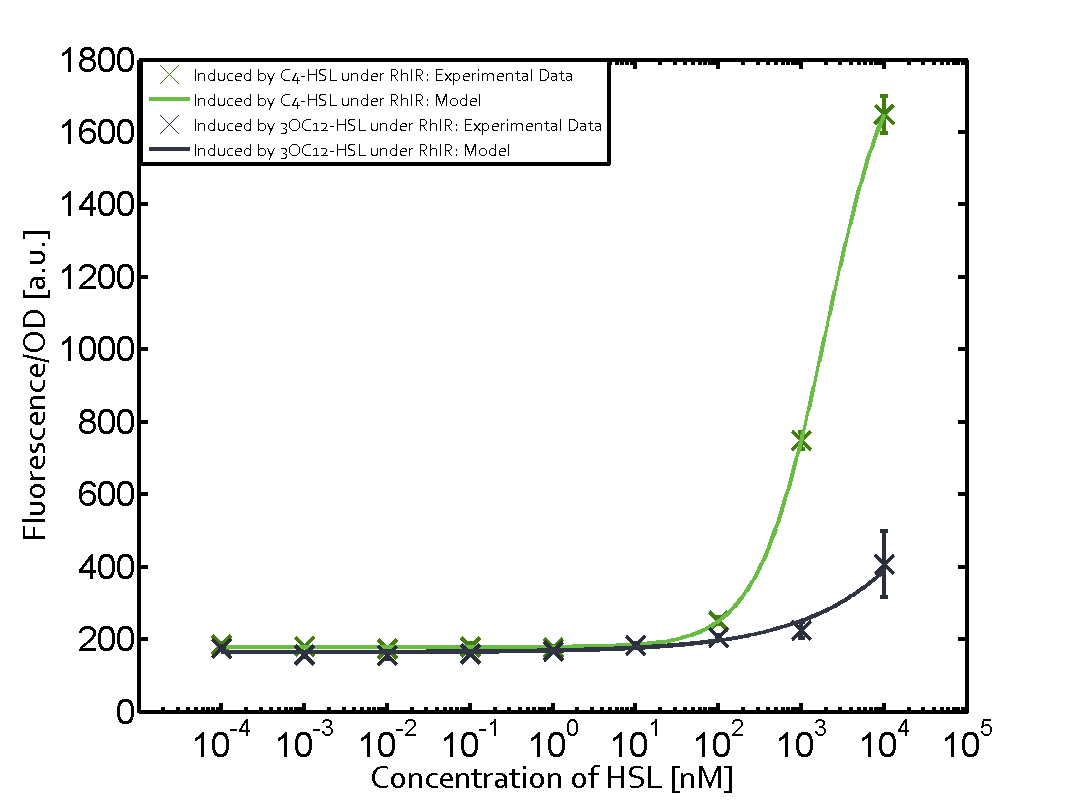
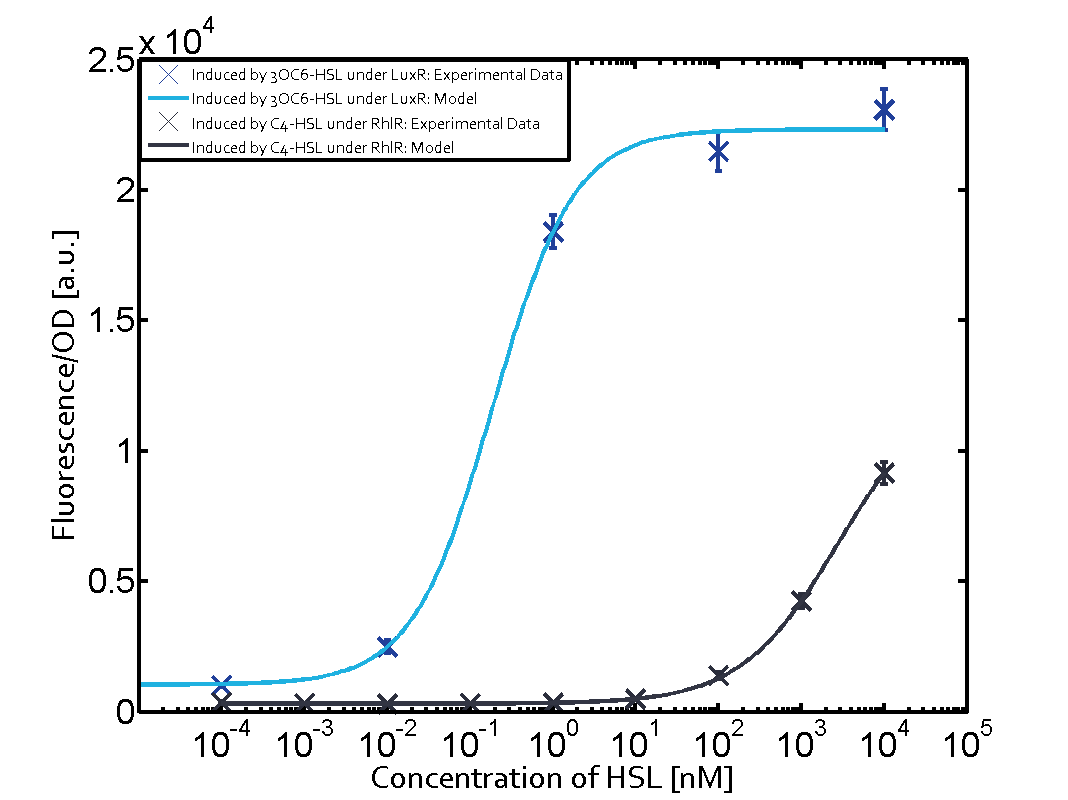
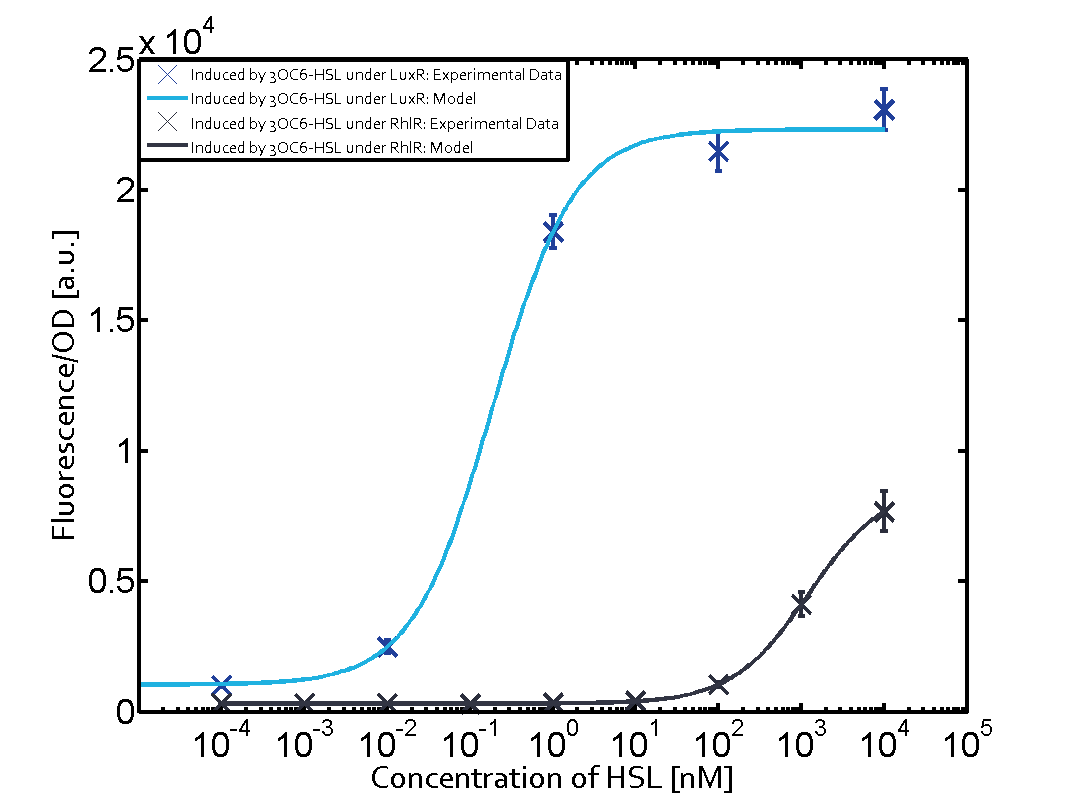
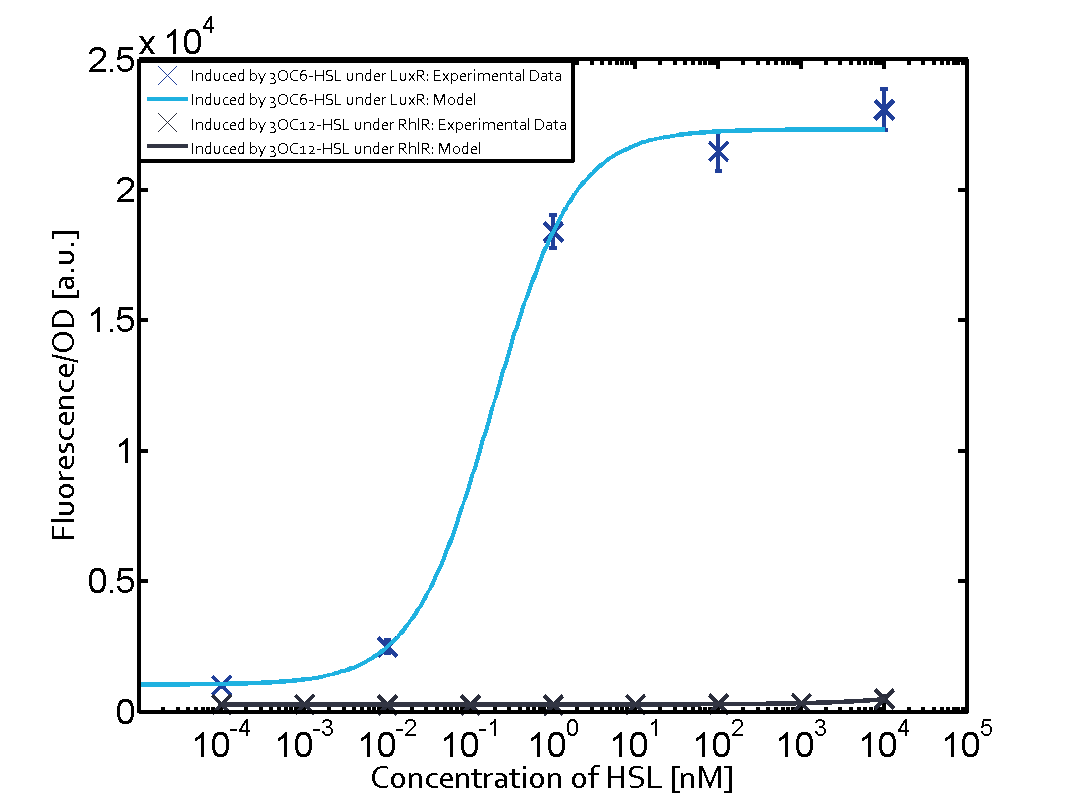
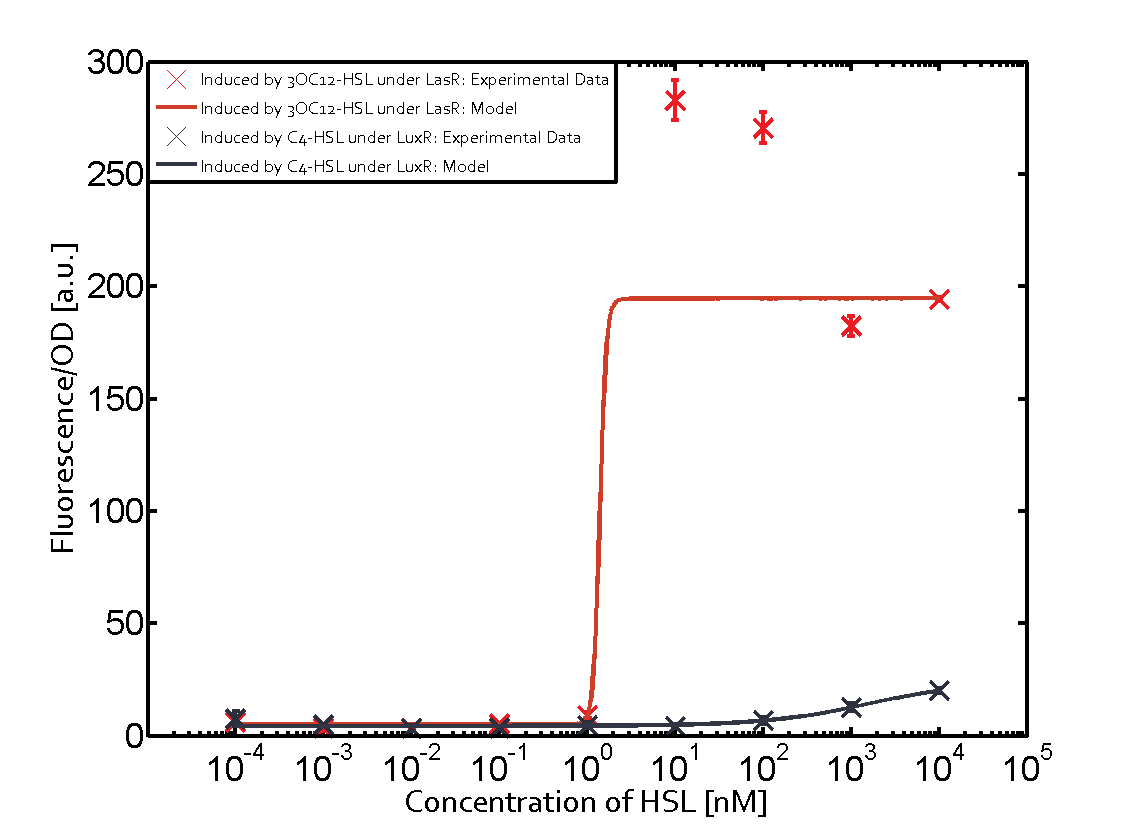
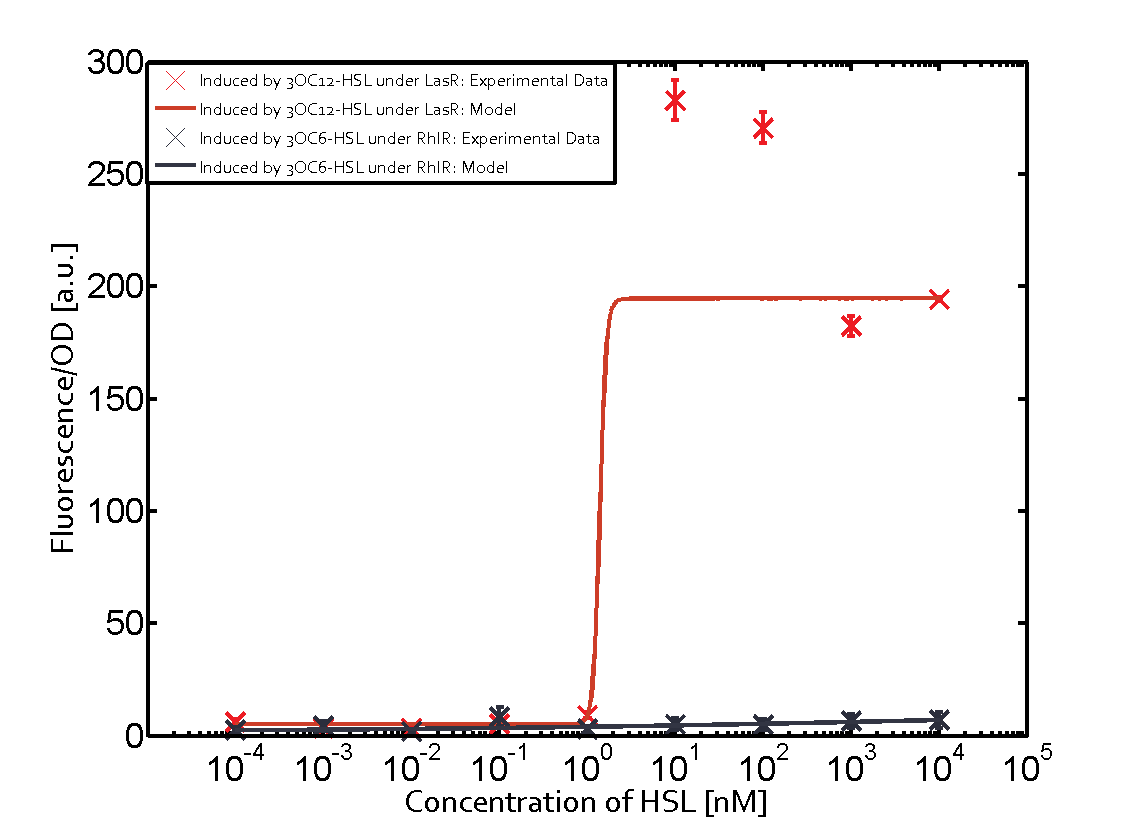
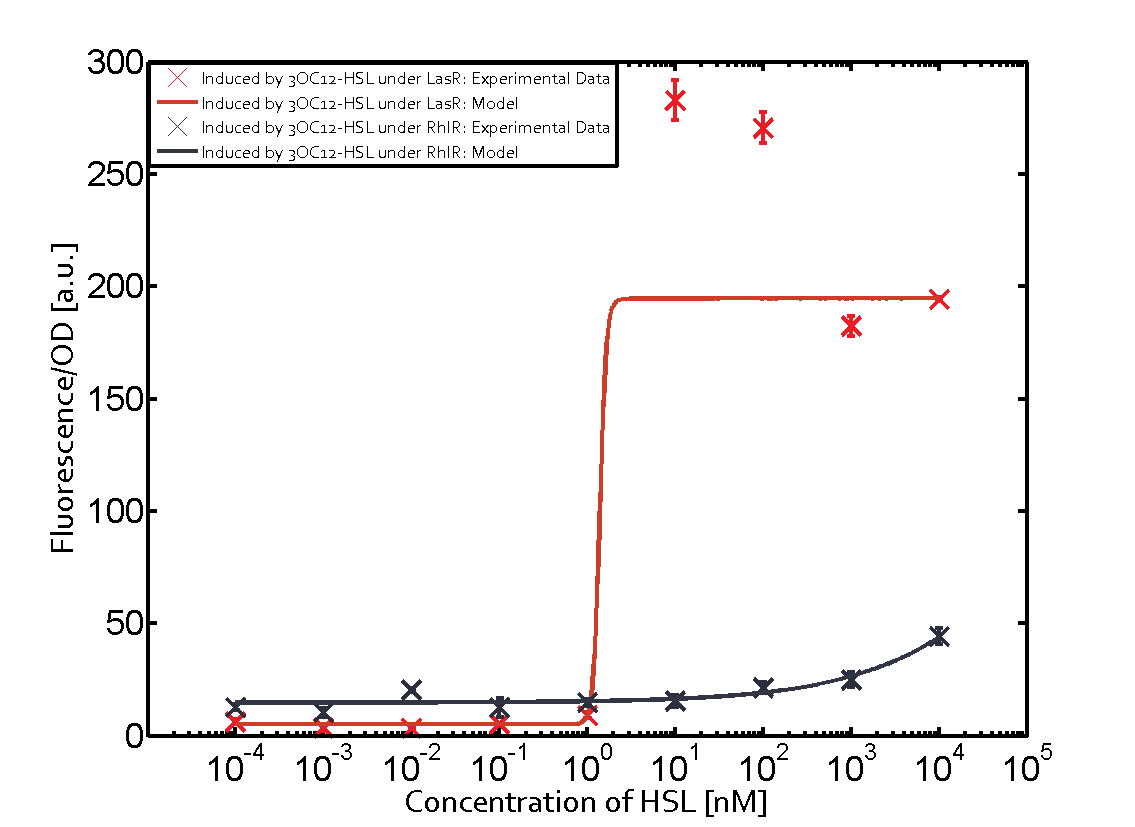
<p>The fitting of the graphs was performed using the following equation :<br><br> | <p>The fitting of the graphs was performed using the following equation :<br><br> | ||
| − | rFluo = the relative fluorescence (absolute measured fluorescence value over OD)[ | + | rFluo = the relative fluorescence (absolute measured fluorescence value over OD)[a.u.]<br> |
| − | a = basal expression rate [ | + | a = basal expression rate [a.u.](“leakiness”)<br> |
| − | b = maximum expression rate [ | + | b = maximum expression rate [a.u.]("full induction")<br> |
n = Hill coefficient (“cooperativity”)<br> | n = Hill coefficient (“cooperativity”)<br> | ||
K<sub>m</sub> = Half-maximal effective concentration (“sensitivity”)<br> | K<sub>m</sub> = Half-maximal effective concentration (“sensitivity”)<br> | ||
| Line 53: | Line 53: | ||
|- | |- | ||
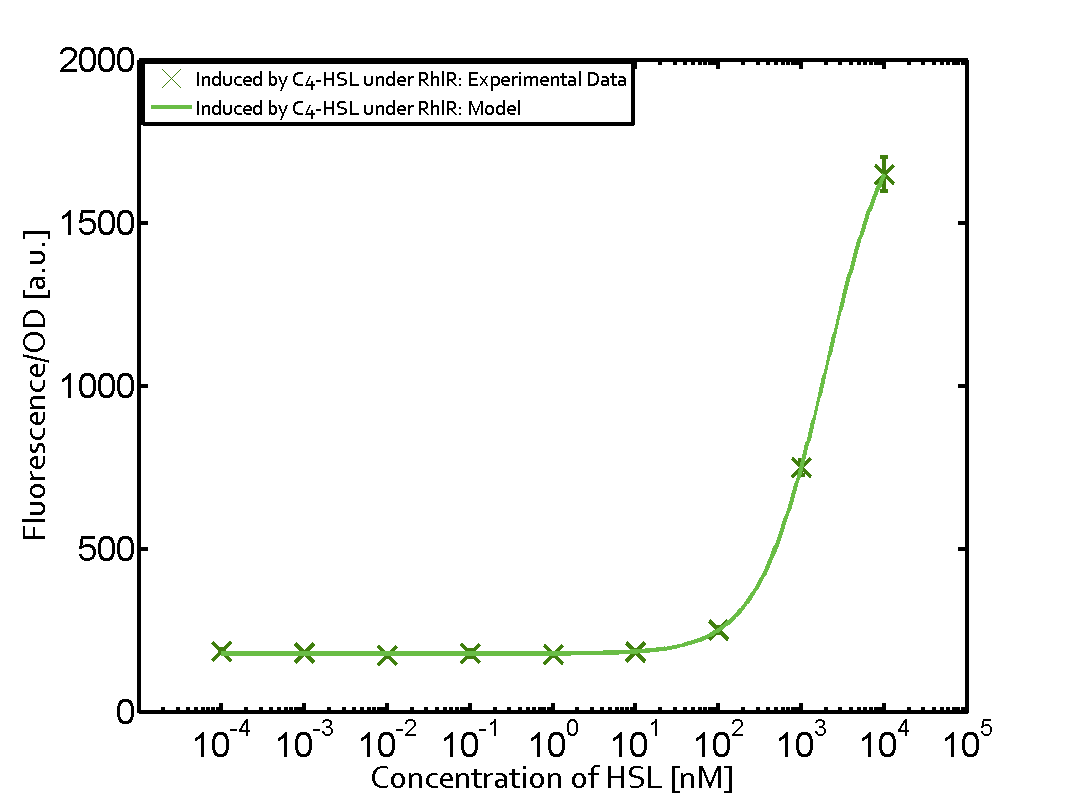
! [https://parts.igem.org/Part:BBa_I14017 Prhl] | ! [https://parts.igem.org/Part:BBa_I14017 Prhl] | ||
| − | | a = 178.4 (174.9, 182) [ | + | | a = 178.4 (174.9, 182) [a.u.] <br> n = 1.053 (0.9489, 1.157)<br> Km = 1969 (1625, 2313) [nM]<br>b = 1736 (1629, 1842) [a.u.]<br> |
| − | | a = 169.1 (155.2, 182.9) [ | + | | a = 169.1 (155.2, 182.9) [a.u.]<br> n = 0.507 (0.2303, 0.7837) <br> Km = 1.08e8(0, 2.681e10) [nM]<br> b = 9.708e4 (0, 1.192e7) [a.u.]<br> |
| − | | a = 162.8 (150.4, 175.1) [ | + | | a = 162.8 (150.4, 175.1) [a.u.]<br> n = 0.404 (0, 0.998)<br> Km = 9.627e8 (0, 7.824e11) [nM]<br>b = 2.537e4 (0, 8.109e6) [a.u.]<br> |
|- | |- | ||
! [https://parts.igem.org/Part:BBa_R0062 Plux] | ! [https://parts.igem.org/Part:BBa_R0062 Plux] | ||
Revision as of 12:56, 24 October 2014
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_C0171
User Reviews
UNIQ8ad2d1f08883572e-partinfo-00000000-QINU
|
••••
ETH Zurich 2014 |
Characterization of two-order crosstalk on the promoterBackground informationModeling crosstalkEach experimental data set was fitted to an Hill function using the Least Absolute Residual method. The fitting of the graphs was performed using the following equation :
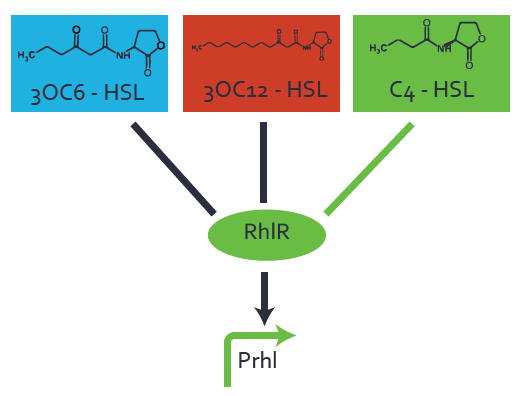
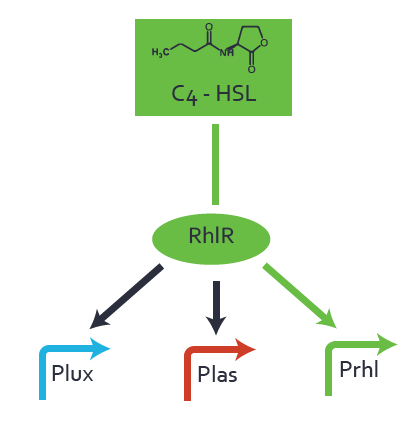
First-order crosstalkFirst Level crosstalk: RhlR binds to different HSL and activates the promoterSecond Level crosstalk: other regulatory proteins, like LuxR, bind to their natural HSL substrate and activates the promoterSecond order crosstalk: Combination of both cross-talk levelsResults
| ||||||||||||||||||||||||||||||||||||
|
Antiquity |
This review comes from the old result system and indicates that this part did not work in some test. |
UNIQ8ad2d1f08883572e-partinfo-00000003-QINU
|
No review score entered. Northwestern 2011 |
The 2011 Northwestern iGEM team used this part as a part of our Pseudomonas Aeruginosa biosensor. We were able to successfully express LasR (C0171) continuously in our system. BBa K575024 |