Part:BBa_K863012
tthl laccase from T. thermophilus with constitutive promoter J23100, RBS and His-tag
tthl laccase (T. thermophilus) with constitutive promoter and HIS tag
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal XbaI site found at 37
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 1436
- 23INCOMPATIBLE WITH RFC[23]Illegal XbaI site found at 37
- 25INCOMPATIBLE WITH RFC[25]Illegal XbaI site found at 37
Illegal NgoMIV site found at 503
Illegal NgoMIV site found at 990 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 818
Initially some trials of shaking flask cultivations were made with different parameters to identify the best conditions for the production of the His-tagged laccase LttH from [http://www.dsmz.de/catalogues/details/culture/DSM-7039.html?tx_dsmzreso Thermus thermophilus HB27] named TTHL. Due to the absence of enzyme activity of the enzyme in the cell lysate a purification method was established (using Ni-NTA-His tag resin). Using E. coli KRX containing BioBrick BBa_K863010, TTHL could not be detected by SDS-PAGE (molecular weight of 53 kDa) or by activity test. Therefore a new BioBrick BBa_K863012 was constructed and expressed in E. coli Rosetta-Gami 2. With this expression system the TTHL could be detected by SDS-PAGE and purified by using a small scale Ni-NTA column. The fractionated samples were tested regarding their activity. TTHL was shown to oxidize ABTS. After measuring activity of TTHL a scale up of the fermentation was successfully implemented up to 6 L.
Cultivation, Purification and SDS-PAGE
Shaking Flask Cultivation
The first trials to produce the LttH-laccase from [http://www.dsmz.de/catalogues/details/culture/DSM-7039.html?tx_dsmzresources_pi5 Thermo thermophilus HB27] (named TTHL) were performed in shaking flasks with various volumes (from 100 mL up to 1 L flasks, with and without baffles) and under different cultivation conditions. The best cultivation condition for BBa_K863010 expressed in E. coli was screened by varying the temperature, the chloramphenicol concentration,induction strategy and cultivation time. Furthermore, E. coli was cultivated with and without 0.25 mM CuCl2 in the medium to provide a sufficient amount of copper, which is needed for bilding the active center. Under the screened conditions no biological active TTHL could be produced. Therefore another BioBrick was constructed and another chassi was chosen. To improve the expression another BioBrick BBa_K863012 was used, which has a constitutive promoter instead of the T7 promoter system. Additionally, the strain E. coli Rosetta-Gami 2 was chosen, because of its ability to translate rare codons. TTHL was then produced under the following conditions:
- flask design: shaking flask without baffles
- medium: [http://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#LB_medium LB-Medium]
- antibiotics: 60 µg mL-1 chloramphenicol and 300 µg mL-1 ampicillin
- temperature: 37 °C
- cultivation time: 24 h
The reproducibility of the measured data and results were investigated for the shaking flask cultivation, but not yet for the bioreactor cultivation.
Fermentation of E. coli KRX with BBa_K863012
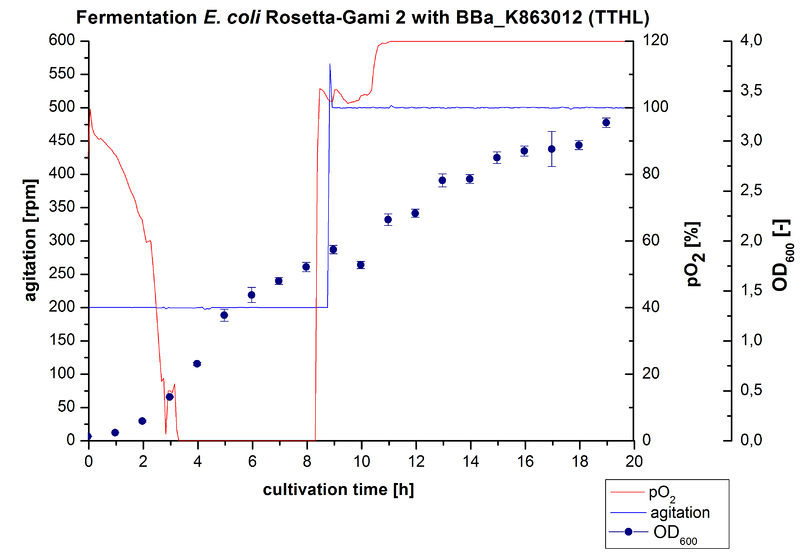
After measuring activity of TTHL we made a scale-up and cultivated E. coli Rosetta-Gami 2 expressing BBa_K863000 in a Bioengineering NFL22 fermenter with a total volume of 6 L. Agitation speed, pO2 and OD600 were online monitored and are illustrated in Figure 1. No initial lag phase was noticeable. Due to the cell growth the pO2 decreased,breakdown of the control unit resulted in a drop to 0%. After a cultivation time of 9 hours the agitation speed was therefore increased manually up to 500 rpm, which resulted in a higher pO2 value of more than 100 % for the rest of the cultivation. During the whole process the OD600 increased slower compared to the fermentation of E. coli KRX expressing BBa_K863000 or BBa_K863005. The maximal OD600 was reached after 19 hours cultivation time at which point the cells were harvested.
Purification of TTHL
The cells were harvested by centrifugation and resuspended in [http://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography Ni-NTA-equilibrationbuffer], mechanically disrupted by [http://2012.igem.org/Team:Bielefeld-Germany/Protocols/Production#Mechanical_lysis_of_the_.28bio-reactor.29_cultivation high pressure homogenization] and centrifuged. After preparing the cell paste the TTHL could not be purified with the 15 mL column, due to a not available column. For this reason a small scale purification (6 mL) of the supernatant of the homogenisation was made with a 1 mL Ni-NTA-column.
SDS-PAGE of purified TTHL
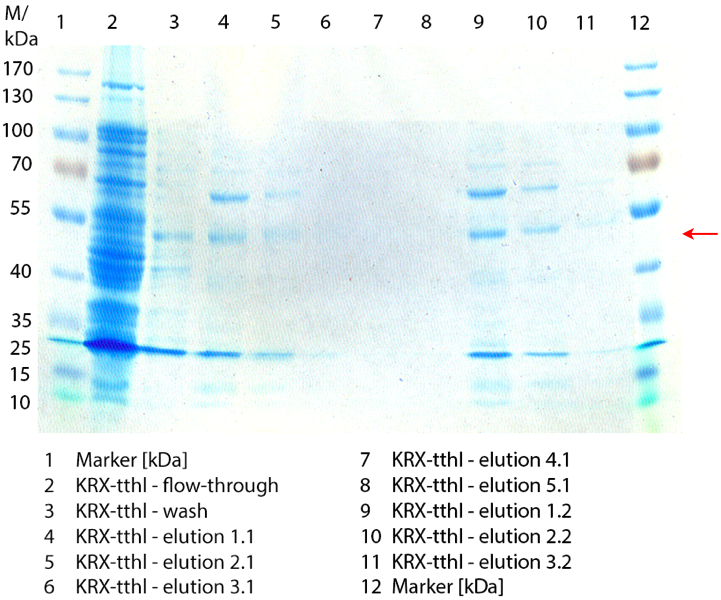
Figure 2 shows the SDS-PAGE of the purified E. coli Rosetta-Gami 2 lysates fermented in 6 L Bioengineering NFL22 fermenter. Additionally the flow-through, wash and all elution fractions (1 to 5) are shown. TTHL has a molecular weight of 53 kDa and the corresponding band is marked with a red arrow. The TTHL band can be found in fractions 1 to 3, but not in the other two elution fractions. Furthermore there are some other non-specific bands, which could not be identified. To improve the purification an 15 mL column was implemented.
Activity analysis of TTHL
Substrate Analysis
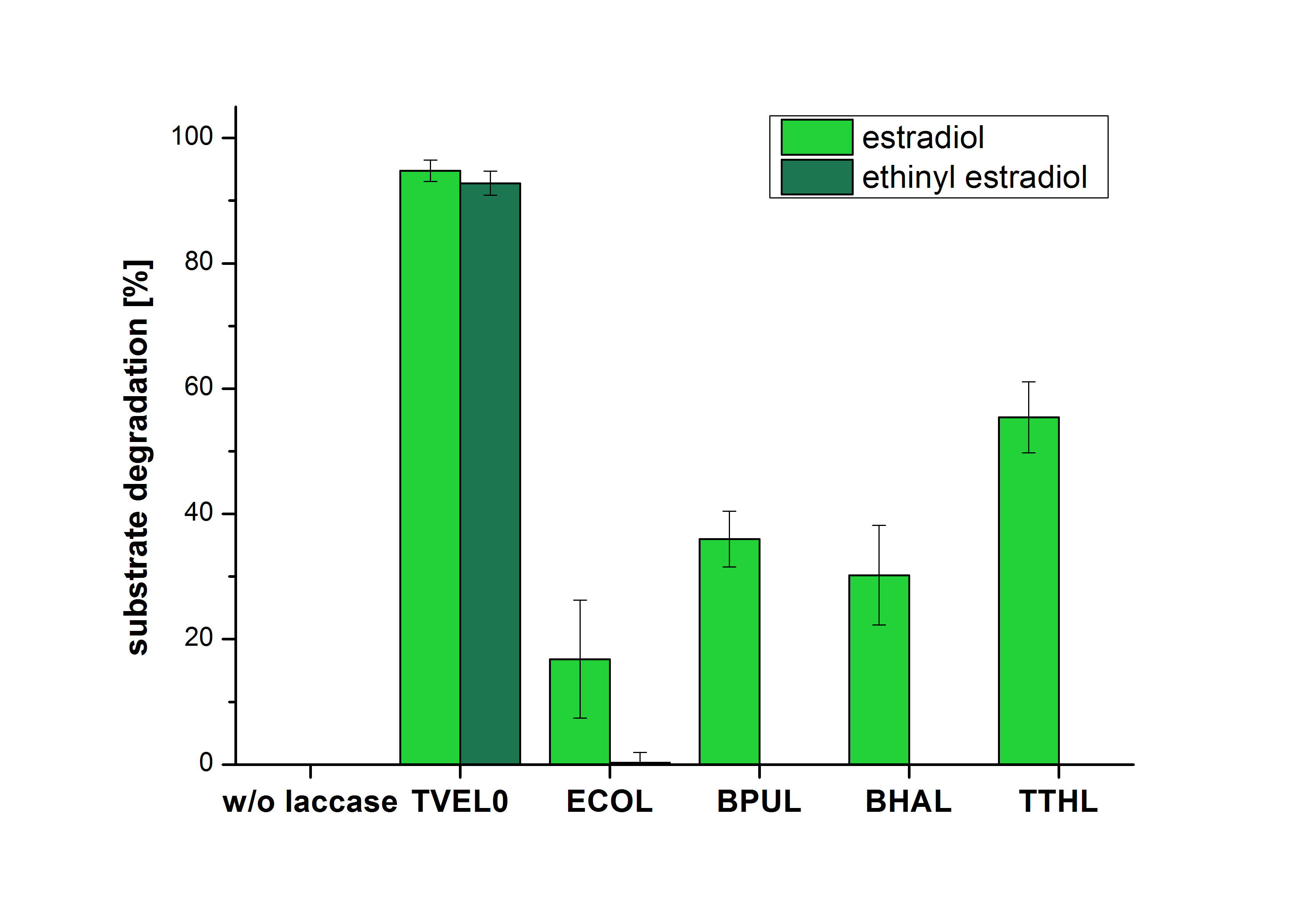
The measurements were made to test if the produced laccases were able to degrade different hormones. Therefore the produced laccases were inserted in the same concentrations (3 µg mL-1) to the different measurement approaches. To work with the correct pH value (which were measured by the Team Activity Test) Britton Robinson buffer at pH 5 was used for all measurements. The initial substrate concentration was 5 µg mL-1. The results of the reactions without ABTS are shown in Figure 2. On the Y-axis the percentages of degraded estradiol (blue) and ethinyl estradiol (red) are indicated. The X-axis displays the different tested laccases. The degradation was measured at t0 and after five hours of incubation at 30 °C. The negative control was the substrate in Britton Robinson buffer and showed no degradation of the substrates. The bought laccase TVEL0 which is used as positive control is able to degrade 94.7 % estradiol and 92.7 % ethinyl estradiol. The laccase BPUL (from Bacillus pumilus) degraded 35.9 % of used estradiol after five hours. ECOL was able to degrade 16.8 % estradiol. BHAL degraded 30.2 % estradiol. The best results were determined with TTHL (laccase from Thermus thermophilus). Here the percentage of degradation amounted 55.4 %.
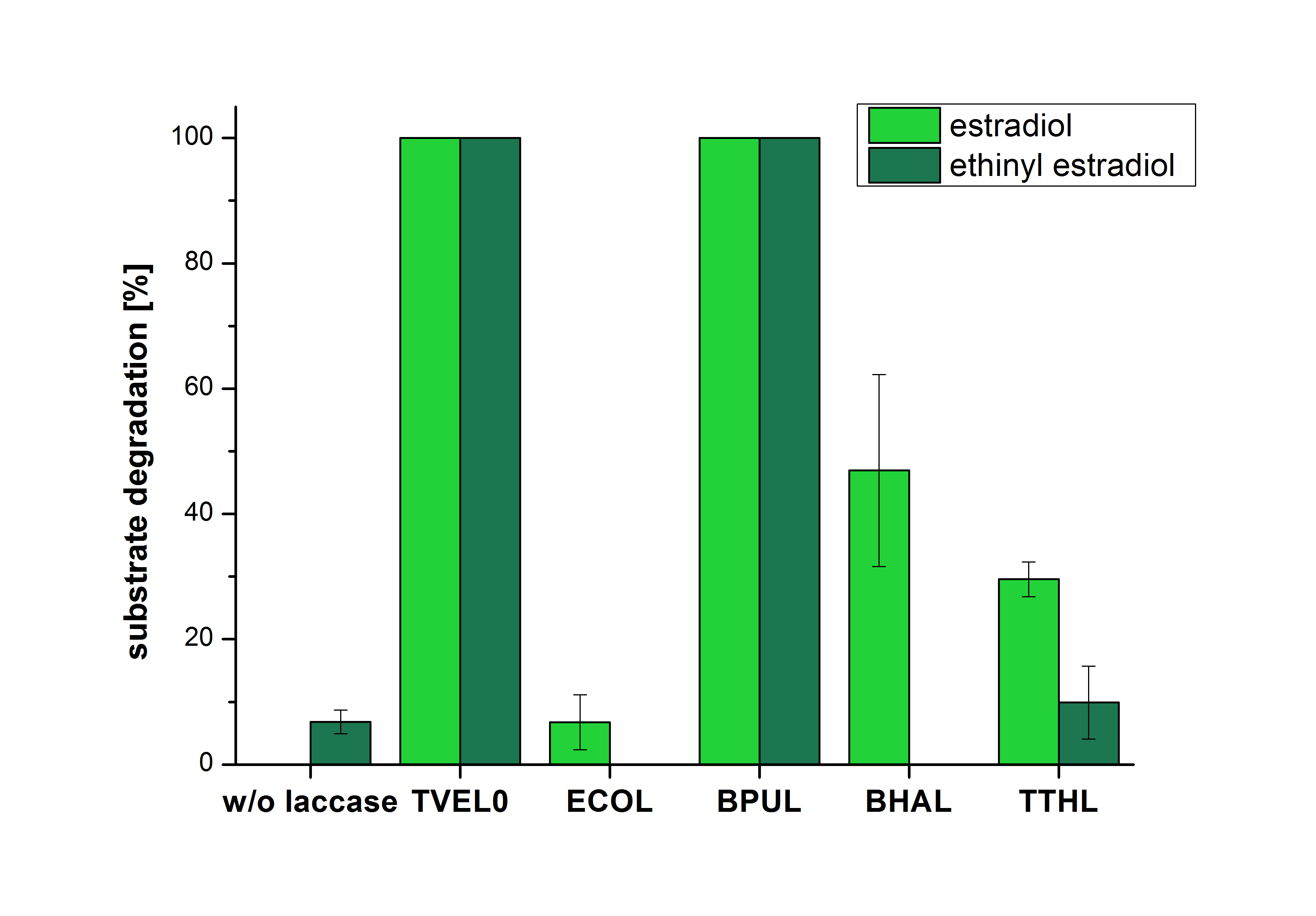
The results of the reactions of the laccases with addition of ABTS are shown in Figure 3. The experimental set ups were the same as the reaction approach without ABTS described above. The X-axis displays the different tested laccases. On the Y-axis the percentages of degraded estradiol (blue) and ethinyl estradiol (red) are shown. The degradation was measured at t0 and after five hours of incubation at 20 °C. The negative control showed no degradation of estradiol. 6.8 % of ethinyl estradiol was decayed. The positive control TVEL0 is able to degrade 100 % estradiol and ethinyl estradiol. The laccase BPUL (from Bacillus pumilus) degraded 46.9 % of used estradiol after ten minutes incubation. ECOL was able to degrade 6.7 % estradiol. BHAL degraded 46.9 % estradiol. With TTHL (laccase from Thermus thermophilus)a degradation 29.5 % were determined.
Immobilization
//function/degradation
//proteindomain/degradation
| None |
