Part:BBa_R0040
TetR repressible promoter
Sequence for pTet inverting regulator. Promoter is constitutively ON and repressed by TetR. TetR repression is inhibited by the addition of tetracycline or its analog, [http://openwetware.org/wiki/ATc aTc].
Usage and Biology
Medium strength promoter. [jb, 5/24/04]
From the reference article:"In contrast to tetracycline, anhydrotetracycline is a particularly useful inducer. It binds Tet R with an ~35-fold higher binding constant and thus allows to operate at very low concentrations. At the same time, its antibiotic activity is ~100-fold lower and concentrations of <50 ng/ml as required for the full induction of P LtetO-1 have no effect on the growth of E.coli."
Modelled by Fudan-CHINA in 2018
This year, Fudan-CHINA makes a mutation to Part:BBa_R0040 to improve its combination with the tTA transcriptional factor by computational simulation.Please be free to visit Part:BBa_K2886010.
Improved by University of Groningen in 2019
University of Groningen has improved the pTet promoter in E. voli using a synthetic promoter library on pTet (BBa_R0040) in order to enhance function and inducibility. Please visit Part:BBa_K3171173 for more info.
Characterized by Fudan in 2019
Background
R0040 is a highly characterized part and many teams before have already characterized it with the concentration-variable inducing curve of its specific inducer aTc and time-lapse inducing curve. The repression effect of ptetR is determined by the expression and combination on particular promotor of tetR, but few teams show their upstream expression level of tetR. This kind of inducing curve lacks of measurement on expression level of upstream tetR. As we use luxpR-tetR-ptetR (R0062-C0040-R0040) for duplex regulation, explicating how the expression level of tetR influence ptetR is essential to us. Therefor, we designed experiments to test ptetR (R0040) under different tetR expression level.
Experience
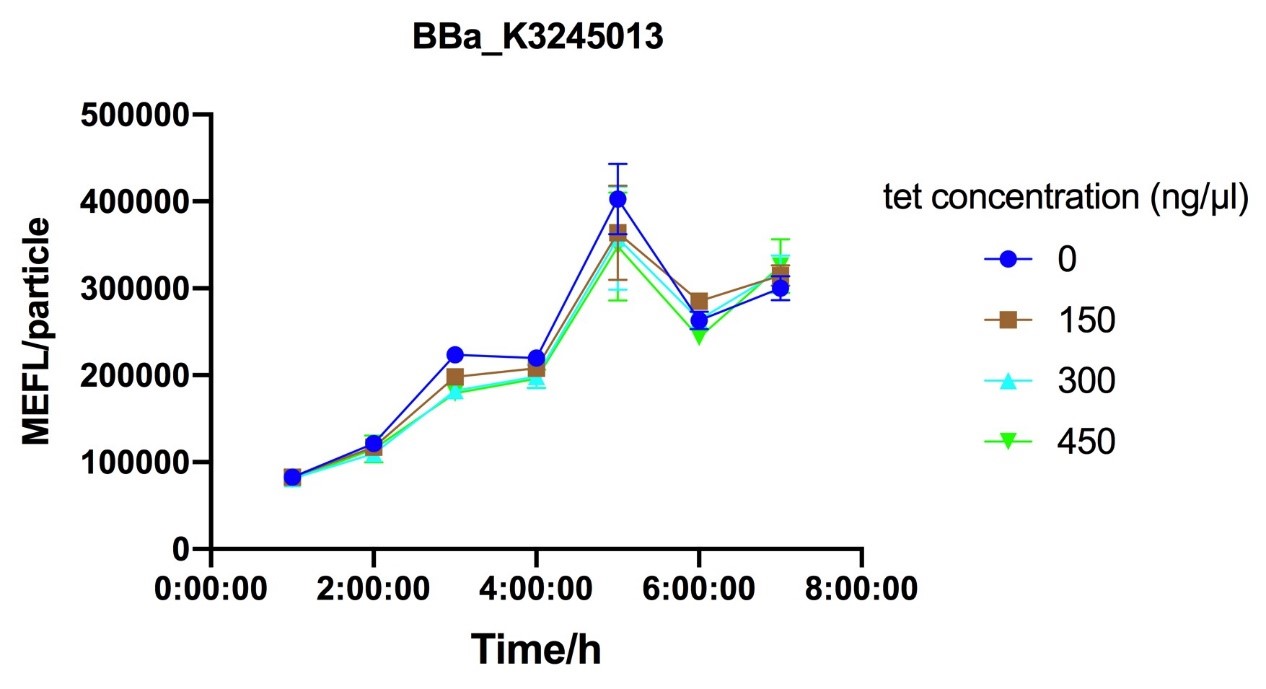
To characterize how R0040 reacts to inducer tetracycline under different concentration of repressor protein tetR, we built 3 parts: BBa_K3245007,BBa_K3245011 and BBa_K3245012, correspondingly using J23106,J23112 and J23115 promotor, which all have been highly characterized, to set up gradient for tetR expression. Since the best inducer aTc is not accessible to us, tetracycline is chosen as an alternate. Besides, a GFP constant expression system, BBa_K3245013, is set as the positive control to find its influence on Nissle growth.
Protocol
We tested BBa_K3245007,BBa_K3245011, BBa_K3245012 and BBa_K3245013 all by the following protocol.
1. Transmit the tested plasmid into DH10B.
2. Inoculate 3 selected single colonies of each kind of plasmid into 3ml LB culture at 37˚C, 220rpm shaking overnight.
3. Pick 33 50ml falcon tubes with 10ml LB culture in each.
4. Dilute the overnight culture in step 2 to 1/500 in falcon tubes in step 3, incubate all to OD=0.5-0.6 (about 2.5 hours).
5. Divide 33 tubes into 3 groups as parallel copies, 11 tubes in each, add tetracycline to the following concentration gradient: 0, 150, 300, 450, 600, 750, 900, 1200, 1500, 1800, 2100. (unit: ng/µl).
6. Culture 33 tubes at 37˚C, 220rpm shaking for 7 hours. During this period, sampling 0.5ml culture in each tube. All samples are centrifuged at 12000rpm, 1 minute. Remove supernatant and add 500µl sterile PBS to resuspend. Take 100µl resuspended liquids into 96-well plate as the final handled sample. Measure the fluorescence at 420nm/528nm and OD600 of samples by 96-well plate reader (100µl as blank).
7. Record all the data in iGEM Data Analysis Template - florescent Standard Curve analysis tables, MEFL/particle are calculated and response curve under tetracycline of different concentration by time is plotted.
Results
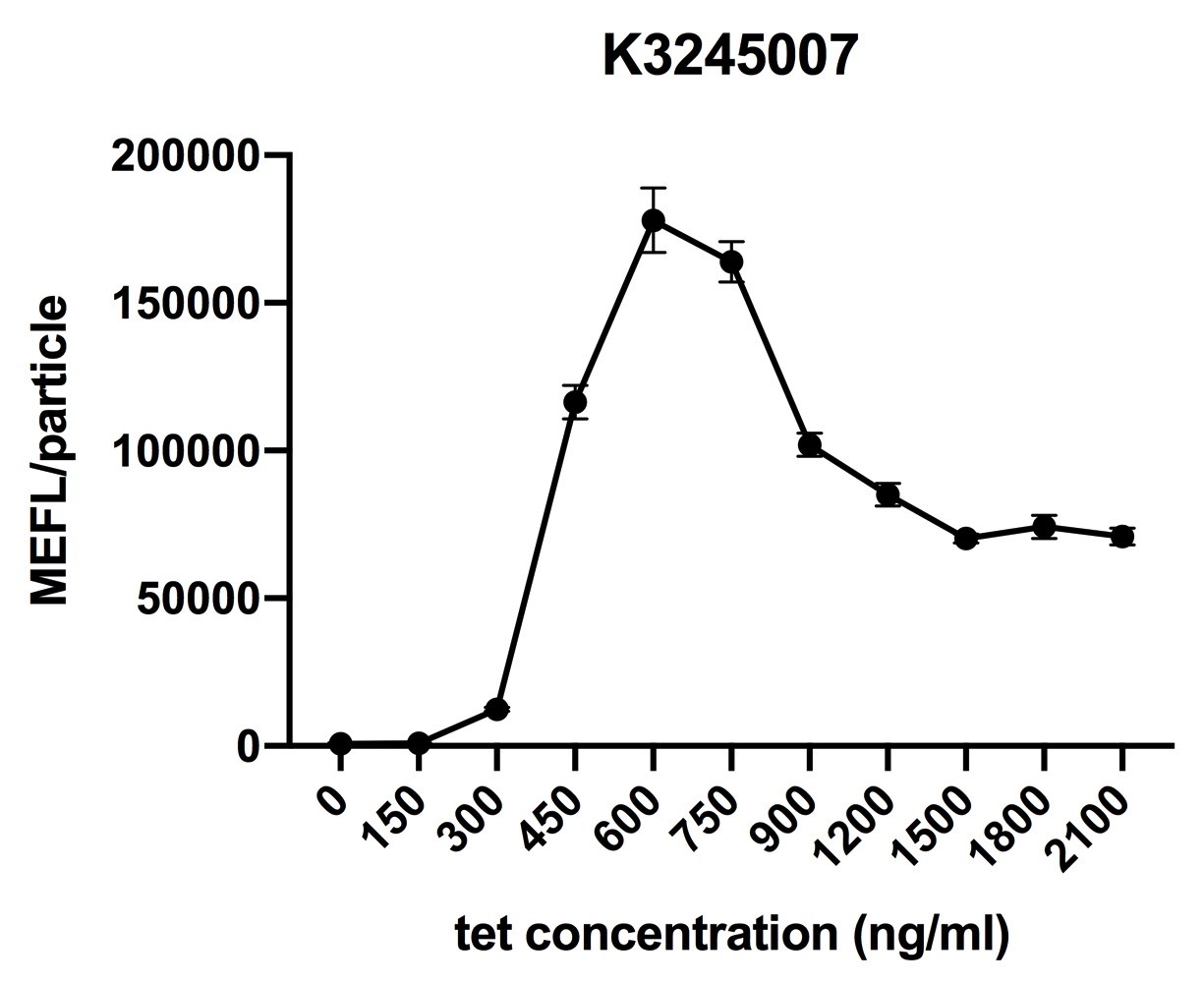
Fig.1 induction curve of ptetR with J23106 upstream expressing tetR (BBa_K3245007) (7 hours). Data are collected and analyzed according to iGEM standard data analysis form after 7 hours of induction.
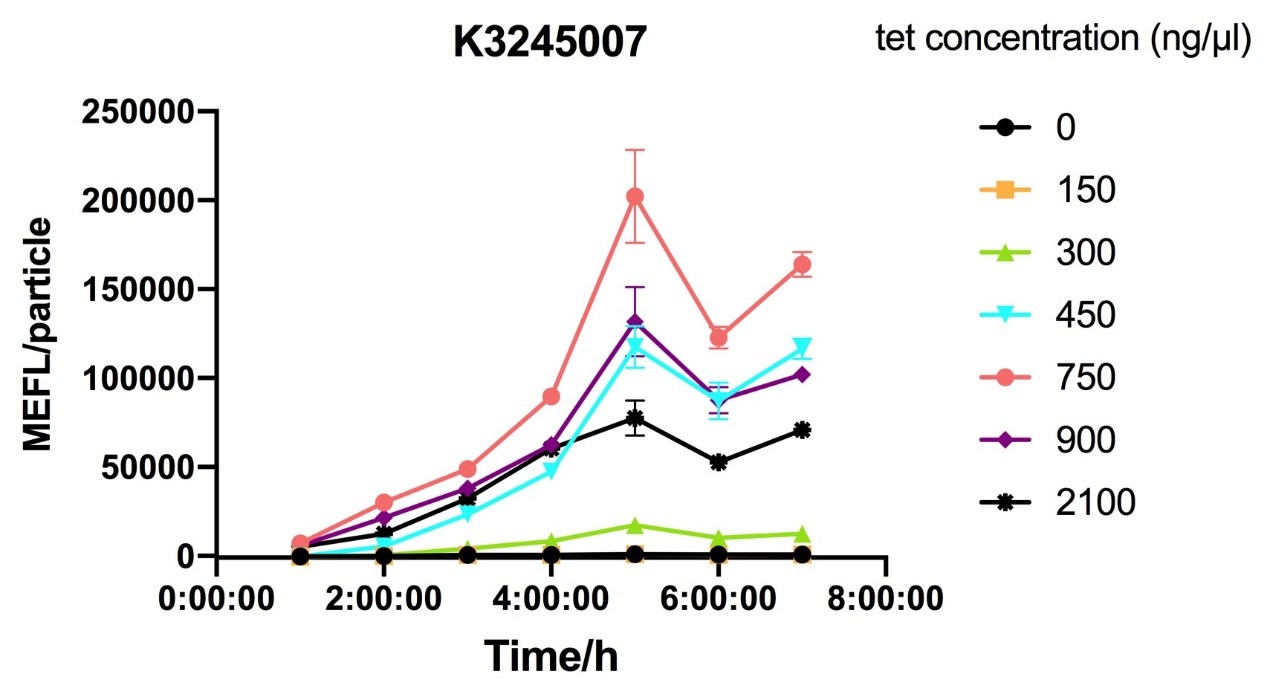
Fig.2 figure showing different induction curves of ptetR with J23106 upstream expressing tetR (BBa_K3245007) under different tet concentration with the lapse of time. Data are collected and analyzed according to iGEM standard data analysis form after 7 hours of induction.
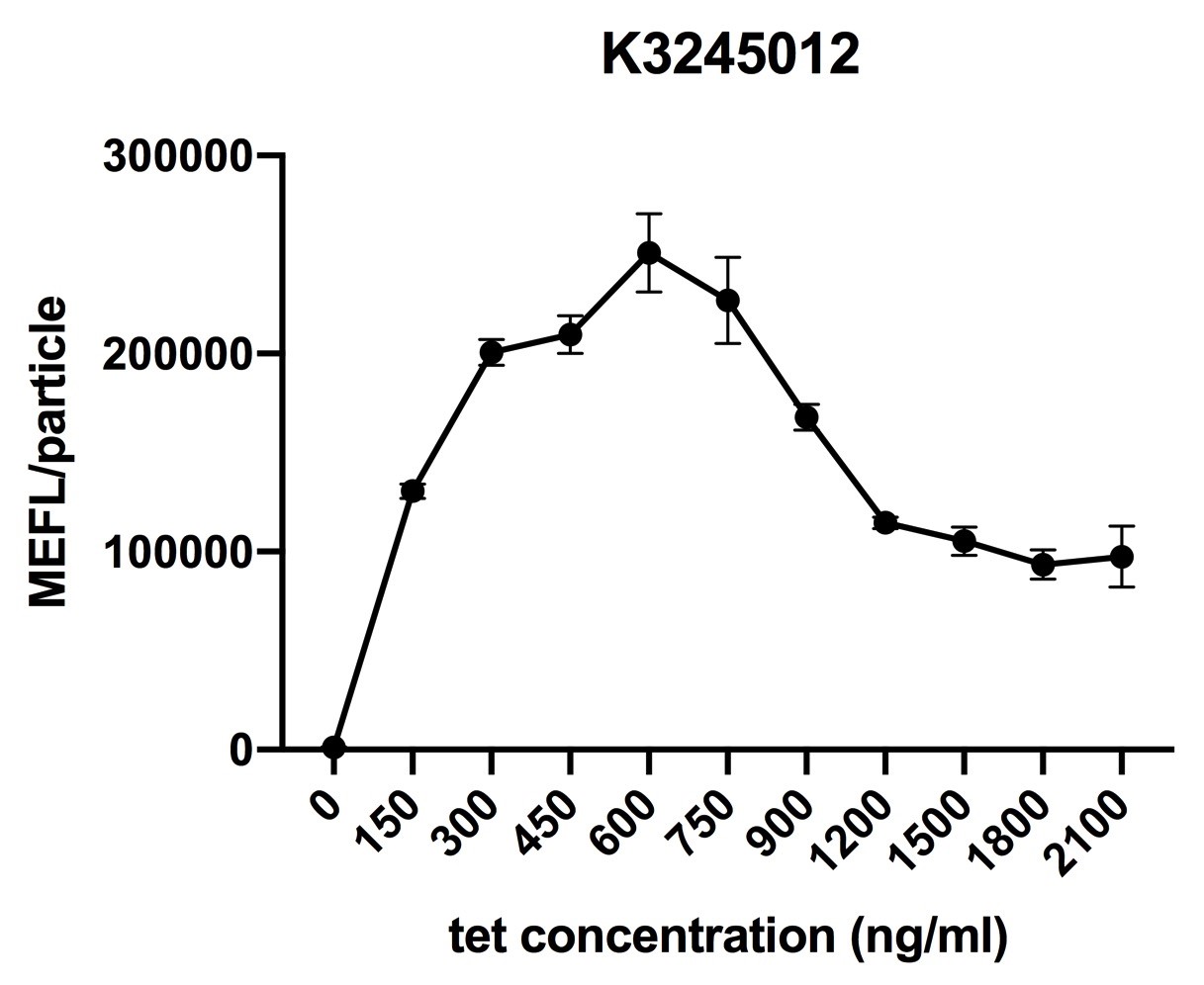
Fig.3 induction curve of ptetR with J23115 upstream expressing tetR (BBa_K3245012)(7 hours). Data are collected and analyzed according to iGEM standard data analysis form after 7 hours of induction.
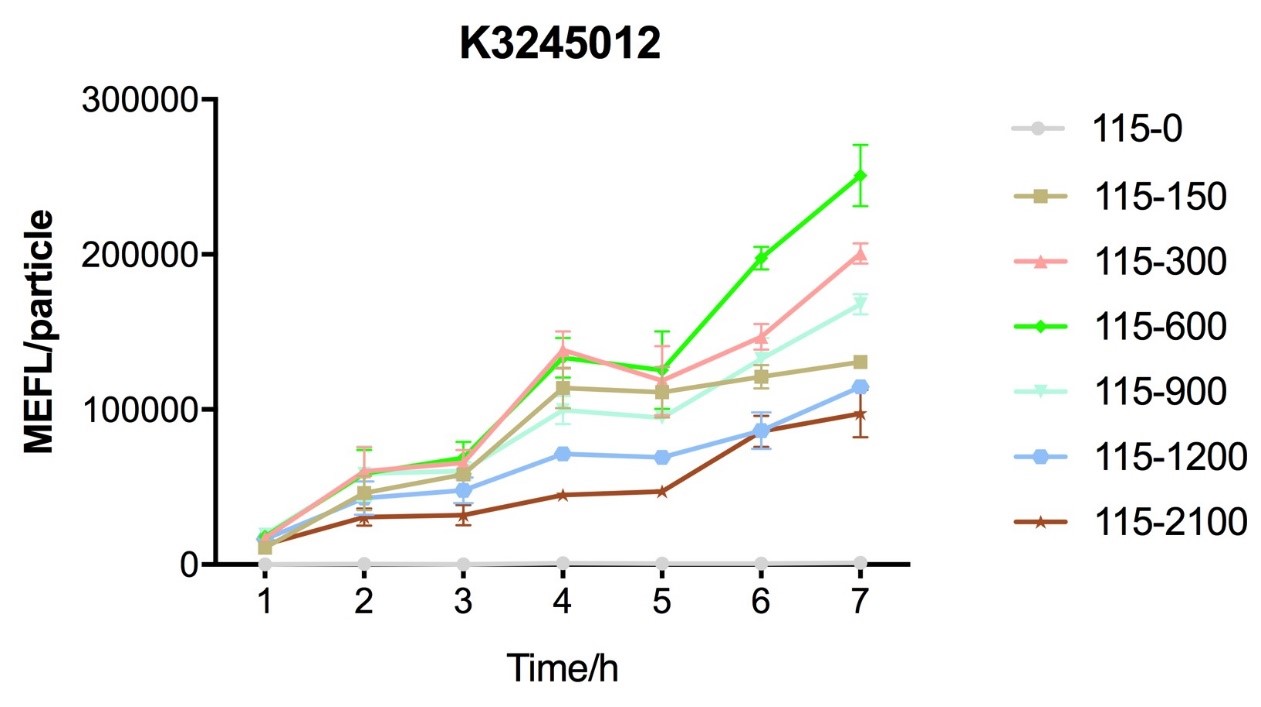
Fig.4 figure showing different induction curves of ptetR with J23115 upstream expressing tetR (BBa_K3245012) under different tet concentration with the lapse of time. Data are collected and analyzed according to iGEM standard data analysis form after 7 hours of induction.
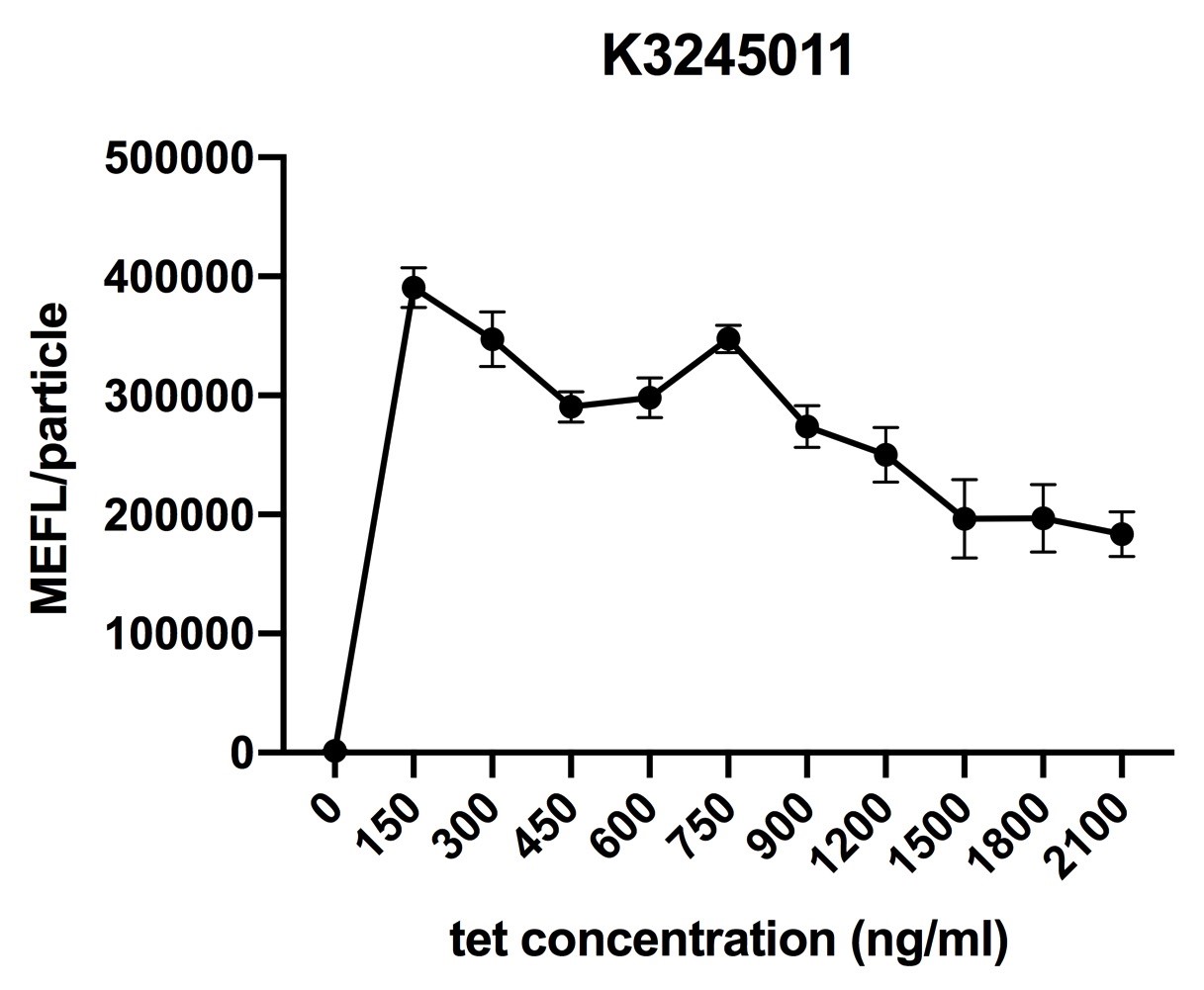
Fig.5 induction curve of ptetR with J23112 upstream expressing tetR (BBa_K3245011) (7 hours). Data are collected and analyzed according to iGEM standard data analysis form after 7 hours of induction.
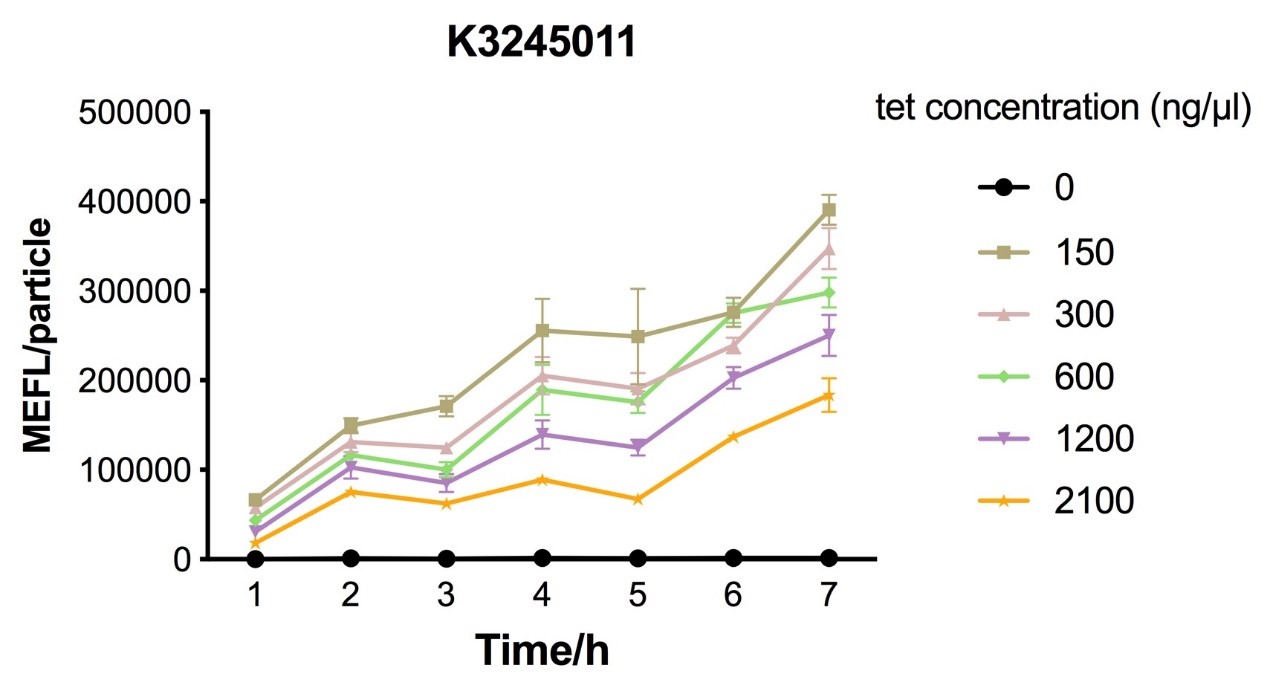
Fig.6 figure showing different induction curves of ptetR with J23112 upstream expressing tetR (BBa_K3245011) under different tet concentration with the lapse of time. Data are collected and analyzed according to iGEM standard data analysis form after 7 hours of induction.
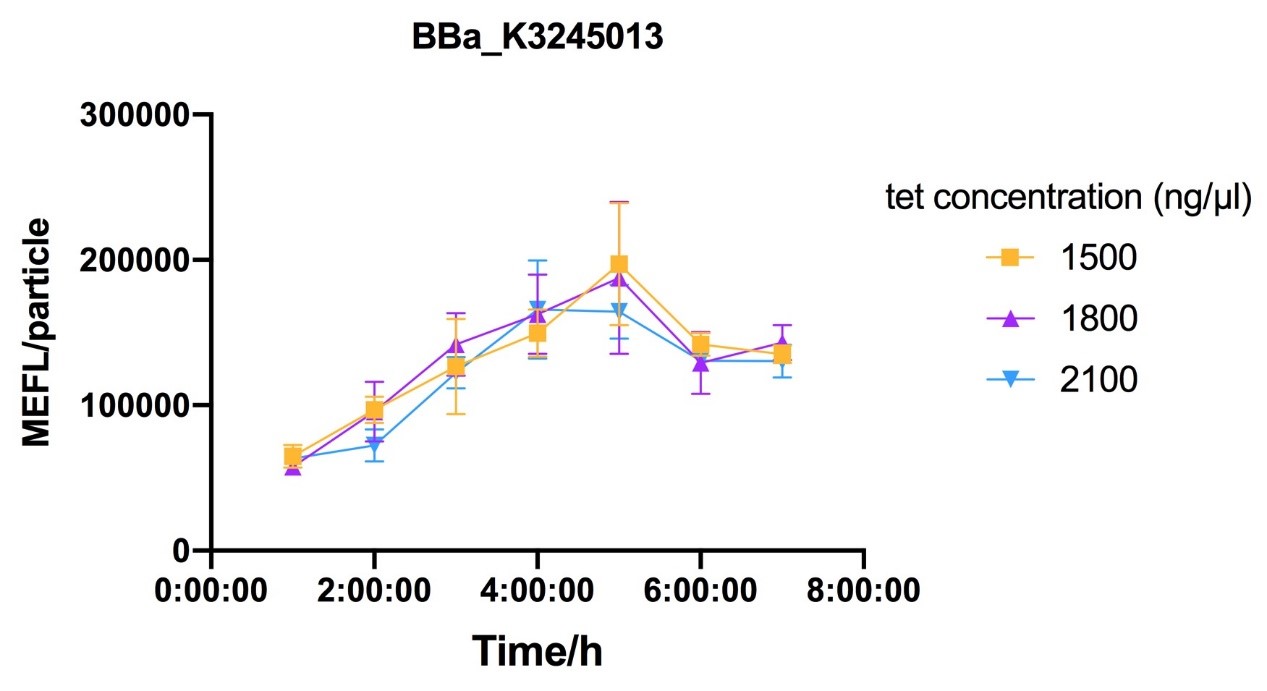
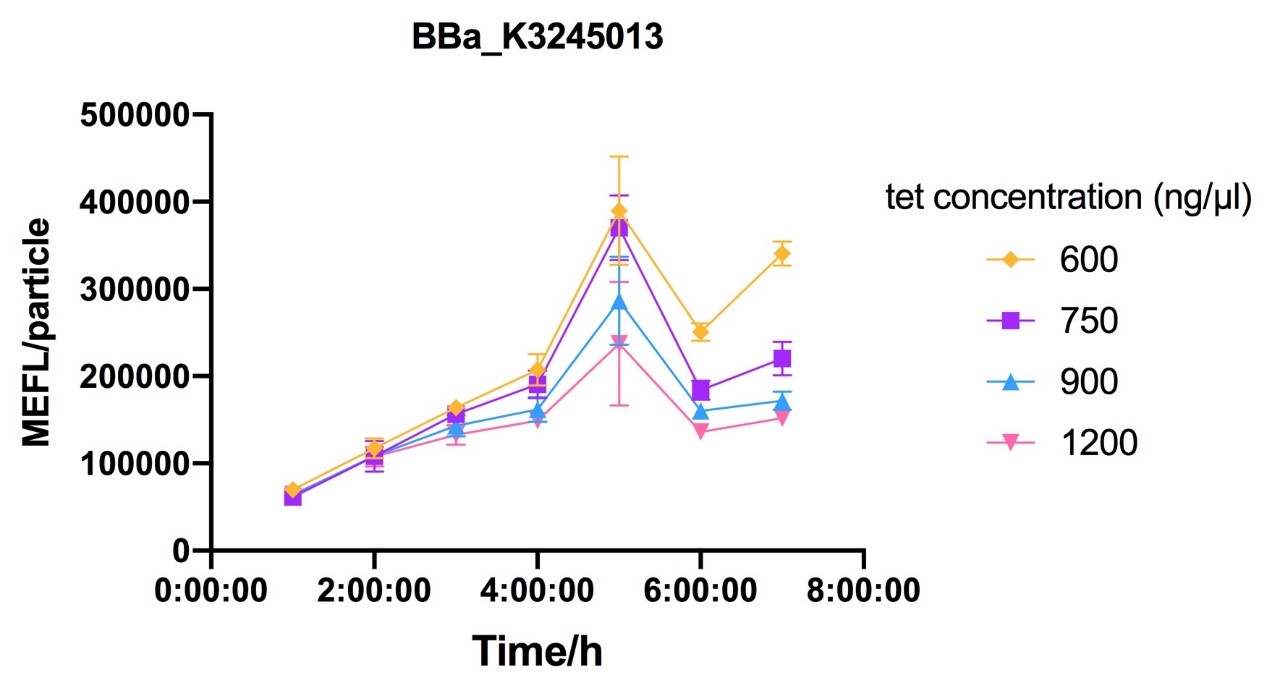
Fig.7-Fig.9 Reaction curves of E. coli DH10B with ptetR-R0034-EsfGFP (BBa_K3245013) against different tet concentration.
Despite the cytotoxic effect of tetracycline on E. coli, we discovered the most suitable tetracycline concentration, 750ng/µl, to induce BBa_K3245007 and BBa_K3245012. MEFL/particle value rises up before 750ng/µl tetracycline with the increase of tetracycline concentration, but appears to steadily descend steadily when passed 750ng/µl threshold. Due to the low level tetR expression in part BBa_K3245011, we discovered a steady descend in MEFL/particle value as tetracycline concentration rises.
Improved by Fudan in 2020
As a well-characterized promoter, PtetR has been studied thoroughly with its expression level and leakage.To better optimize the antimicrobial peptide expressing system, here we take a unique idea to created a new promoter fusion to create a new promoter that are regulated by tetracycline while with a desired expression intensity. It is a fusion of tetO operon and the -35 upstream, -35--10 parts,-10 downstream of the stronger PL promoters in lambda phages, to create a improved promoter with proper expression level while regulated by tetracycline. Please be free to visit Part:BBa_K3606007 and Part:BBa_K3606006.
Usage in non model organisms: Uni-Padua-IT 2023
Acinetobacter baumannii
We attempt the cloning of an expression cassette bearing the pTet promoter driving RFP (final construct, BBa_I13521) inside our plasmid backbone for A. baumannii and K. pneumoniae BBa_K4727000. Once successful cloning was achieved, we tried to electroporate the assembled construct in A. baumannii, using our optimised protocol. After the transformation, some red colonies could be seen; so, a plate reader experiment was carried out to assess the ability of the pTet promoter to allow protein expression in this bacteria.

From the resulting data we can assess that BBa_R0040 can express proteins in A. baumannii at a level significantly lower than the one shown by the same promoter in E. coli, but higher than the weak promoter J23101. The shown expression level could be compared to the one of BBa_J23119 in A. baumannii.
HS_Shanghai 2023 's contribution
Detecting the leakage rate of pTet
To test the leakage efficiency of pTet, HS_Shanghai team constructed two kinds of J23100-B0034-TetR-pTet-B0034-eGFP (+TetR) and pTet-B0034-Egfp (-TetR). To simplify the comparison, we set the fluorescence intensity of NT to 1. The results showed a leakage rate of 2.1% for +TetR.

Figure 1. fluorescence intensity and OD600 control of J23100-B0034-TetR-pTet-B0034-eGFP (+TetR) and pTet-B0034-Egfp (-TetR). Excitation light of 488 nm and emission light of 507 nm, n= 3.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
//direction/forward
//promoter
//regulation/negative
//rnap/prokaryote/ecoli/sigma70
| biology | |
| control | aTc, tetracyline |
| direction | Forward |
| negative_regulators | 1 |
| o_h | |
| o_l | |
| positive_regulators |

 1 Registry Star
1 Registry Star