Part:BBa_I13521
Ptet mRFP
Constitutive on mRFP (positive control)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 635
Illegal AgeI site found at 747 - 1000COMPATIBLE WITH RFC[1000]
Pictures
Part characterization by iGEM Sorbonne Université 2019
DAY 1:
The DNA corresponding to this part is amplified and extracted from liquid cultures of bacteria thanks to the Promega extraction kit.
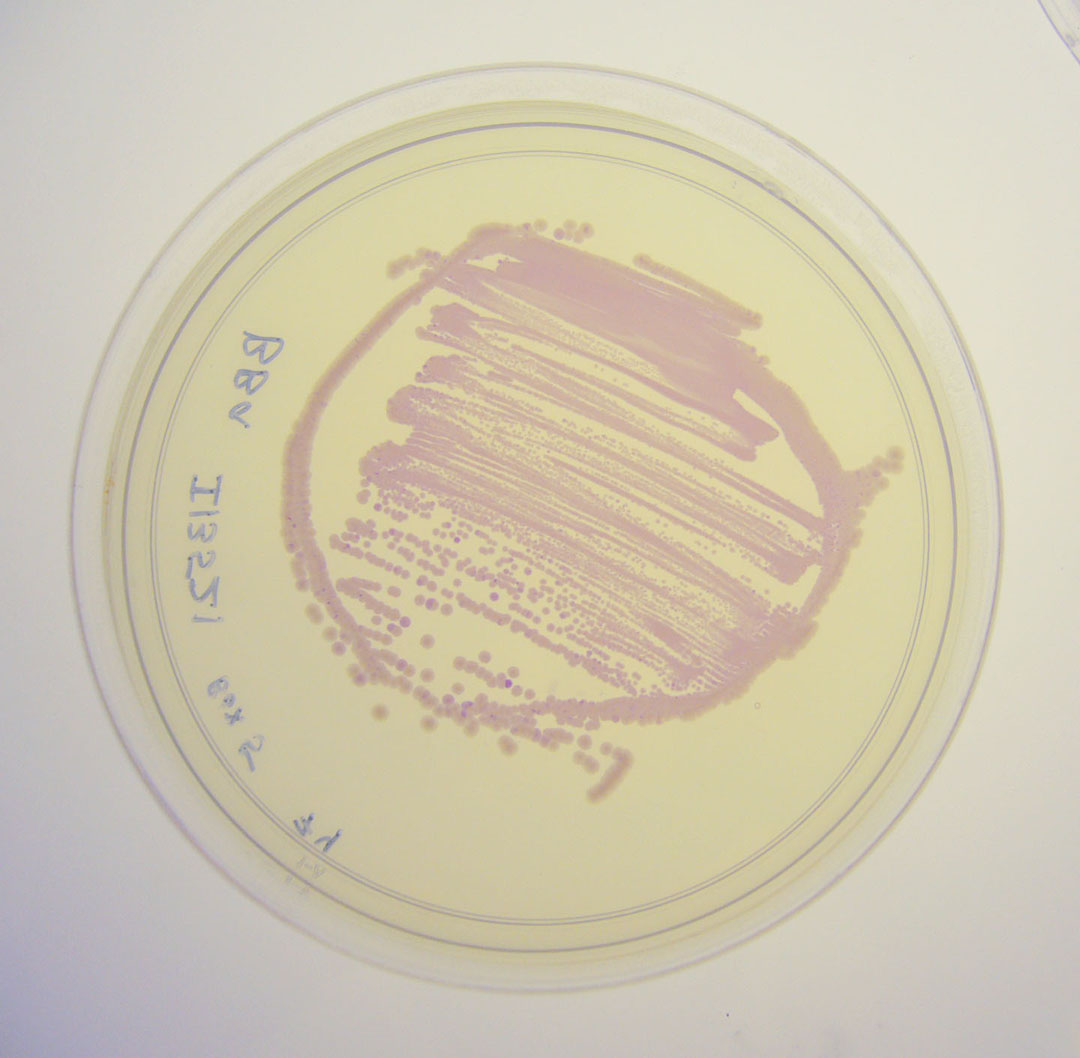
Control digests by Eco RI and Pst I were carried out in order to verify the presence of the part of 923 bp. DNA fragments were analyzed on a 1.2% agarose gel. The 8 clones tested have the correct BBa_I13521.


The plasmid DNAs containing part 1 to 8 are used to transform competent DH5 α bacteria which are then plated on LB medium + Chloramphenicol 34 μg/mL and incubated overnight at 37°C.
DAY 2:
A clone resulting from each transformation is subcultured in liquid culture of 5 mL of LB medium + Chloramphenicol 20 μg/mL. These cultures are incubated overnight at 37°C with 200 rpm shaking.
DAY 3:
The 8 liquid cultures are diluted to an OD at 600 nm of 0.1 in LB medium + Chloramphenicol 20 μg/mL.
The liquid cultures are then placed at 37°C. with stirring at 200 rpm until an OD600 of 0.4 is reached.
Each of the 8 liquid cultures is divided into 3, corresponding to 3 different concentrations of tetracycline: 0 ng/mL - 100 ng/mL – 10 000 ng/mL.
A time course experiments is realized, measuring the OD600 and mRFP fluorescence (535 nm (excitation) / 620 nm (emission) of 100µL of each culture every 30min.
As expected, the more tetracycline there was, the slowly the OD 600 increased, meaning that even at this low concentration, the tetracycline is toxic for bacteria. mRFP fluorescence was too low to overpass background signal (data not shown).
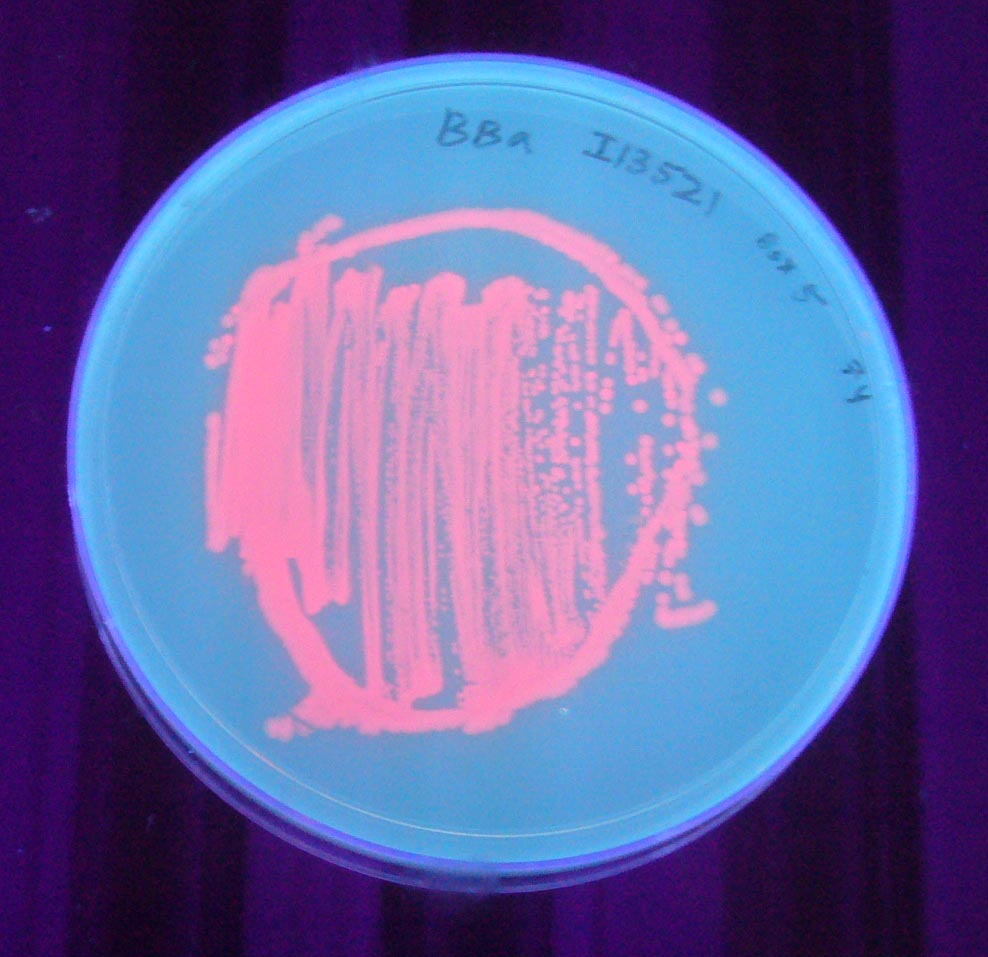
To check if bacteria were expressing the mRFP, we observed the cells after 6h incubation at 37°C using an epi-fluorescence microscope. Doing so, we were able to detect mRFP signal, the bacteria being red. Therefore, we quantified the mRFP fluorescence signal for the bacteria transformed with the BBa_I13521 part.

We observed no significative variation of the mRFP signal depending on the tetracycline concentration.
The BBa_I13521 part is described as “Constitutive on mRFP (positive control)” but the subpart description is not coherent with this description, as it is stated that this reporter construct has a ptetR promoter (regulated by tetracycline, thus not constitutive). The two description being incoherent, we wanted to identify the correct one.
Our data are in favor of a constitutive expression of the mRFP, not affected by the addition of tetracycline.
The followings were edited by iGEM17_SCU-China
Improvement
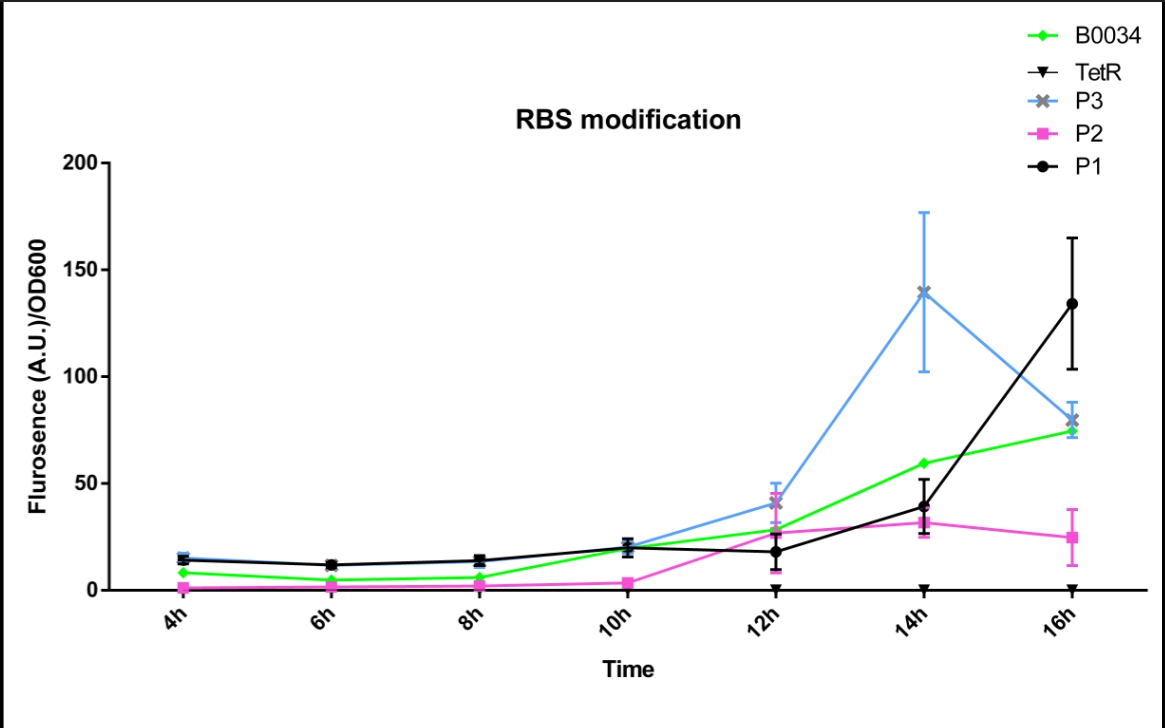
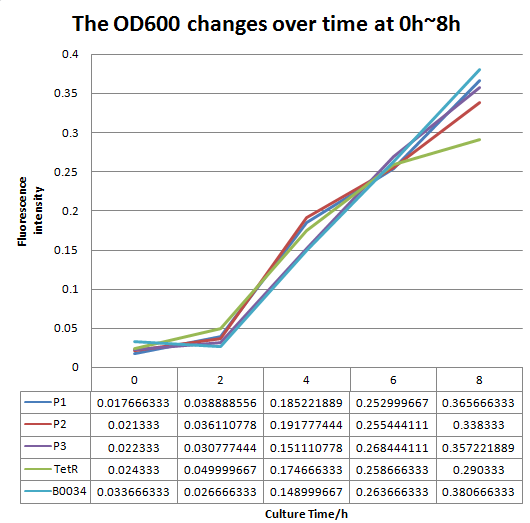
We altered the RBS region and constructed new parts that has higher translation rate, so that the expression of this part, BBa_I13521, could increase, which may benefit the properties of BBa_I13521 as a reporter device with higher sensitivity. To test the performance, we measure the fluorescence intensity and OD600 changes of the bacteria liquid over time within 16hours after inoculated. The result showed that we indeed improved the Part:BBa_I13521 and got a new part, BBa_K2276010, containing a stronger RBS. Fluorescence quenching may explain the drop of the fluorescence intensity of P3 strain at 16h because the concentration of fluorescent protein was too high.
Expression in TetR producing strains - Alma 2020
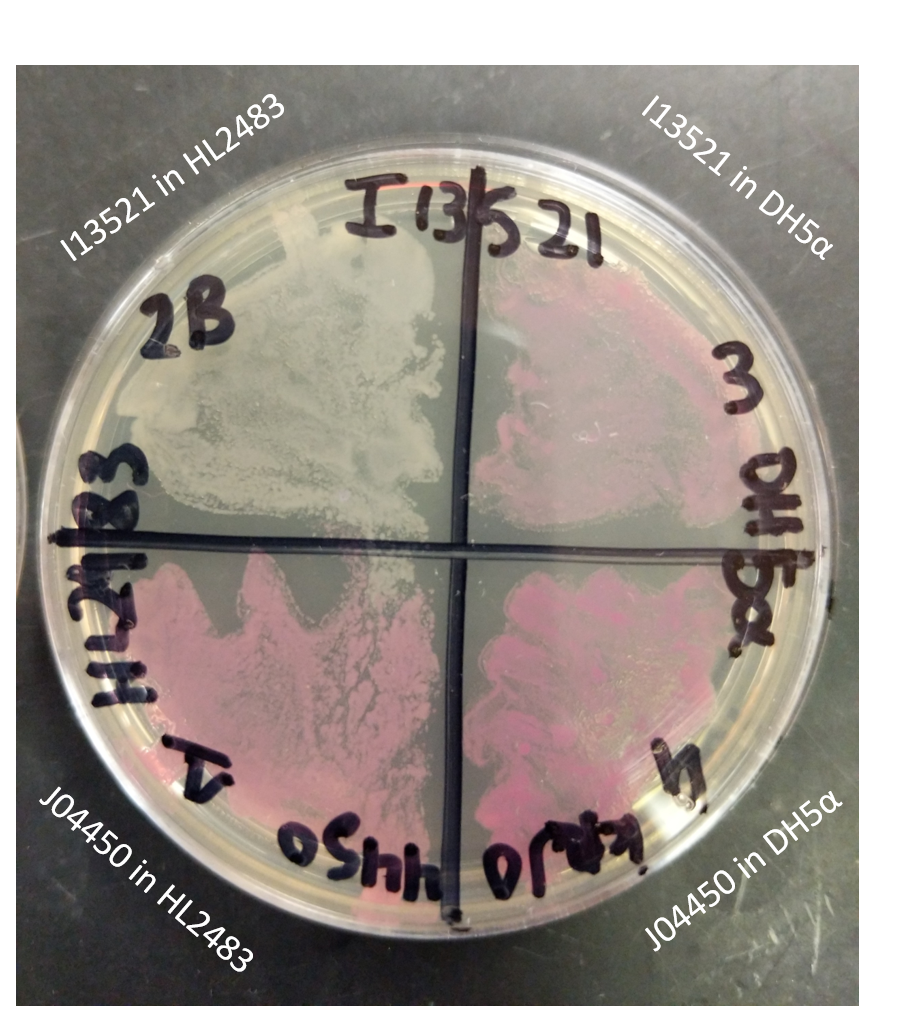
As I13521 is under the control of a TeT repressible promoter we elected to further investigate the control this promoter displays on the output of RFP. In order to do this, we utilized the HL2483 strain of E. coli, which constitutively produces TetR. Therefore, this strain of E. coli would be expected to inhibit the I13521 output of RFP. We tested this strain in conjunction with DH5ɑ E. coli also transformed with I13521. Each strain was additionally transformed with J04450, an RFP producing BioBrick under the control of the LacI promoter, to act as a positive control. These two sets of transformed strains were then streaked onto a LB (Miller) agar plate with chloramphenicol and incubated at 37℃ overnight.
The plate shown below indicates that RFP production was inhibited in the presence of TetR in the HL2483 strain while it was unaffected (and therefore properly functioning) in the DH5ɑ strain. This confirms the control of the RFP output through the TeT repressible promoter.
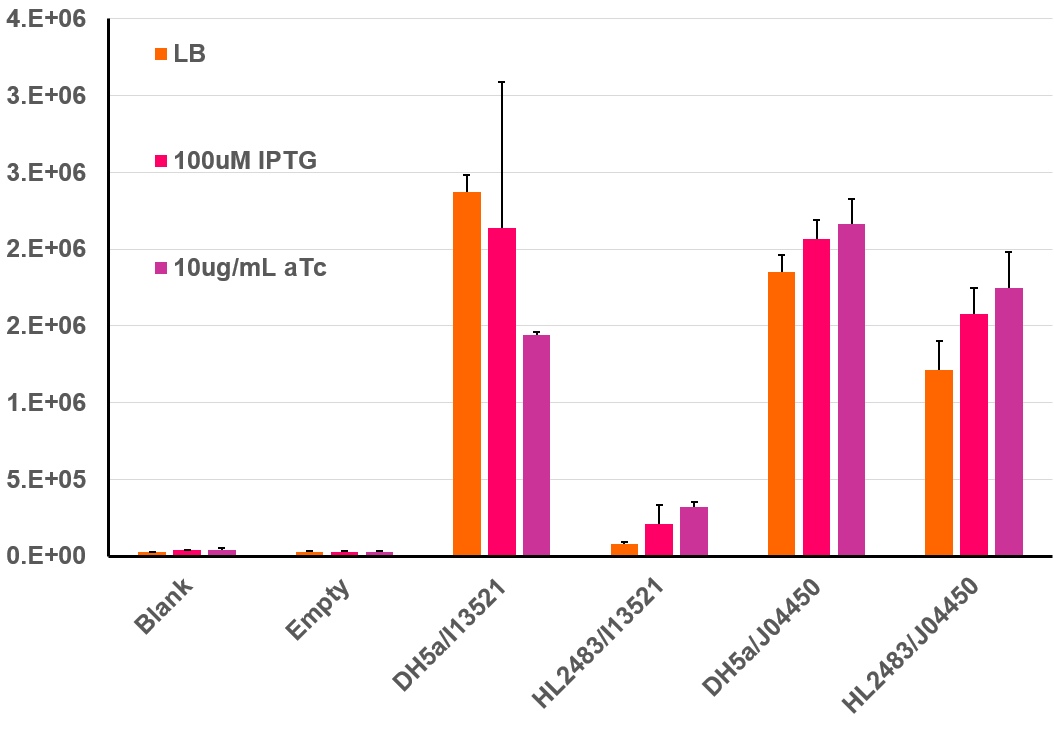
This BioBrick was further characterized by isolating colonies from the above plate and prepping liquid cultures for fluoroscopic analysis. For this experiment, cells were initially prepared according to the 2018 InterLab protocol, but after dilution to a low OD-600, 200uL of each culture were incubated at 37℃ in a 96 well clear bottom black plate. This type of plate allows for both absorbance and fluorescence measurements, and this data was collected every 30 minutes over a 6 hour incubation period. The resulting data can be seen in the bar graph below. This bar graph shows the fluorescent data for the same strains of E. coli discussed above. Additional information was garnered regarding the function of the I13521by adding both IPTG as well as aTc to the plates and measuring the fluorescence compared to cells in LB only. IPTG acts to induce the transcription of genes under control of the Lac promoter. As J04450 is under the control of the LacI promoter, it would be expected to increase its expression in the presence of IPTG in comparison to cells containing only LB, which is what is seen above. Additionally, aTc acts as an inducer to TetR; therefore, in the presence of aTc we would expect an increase in the fluorescence of HL2483 strains with I13521 compared to the LB culture. This result is also seen in the chart below.
Filter disc assay for induction - Alma 2021
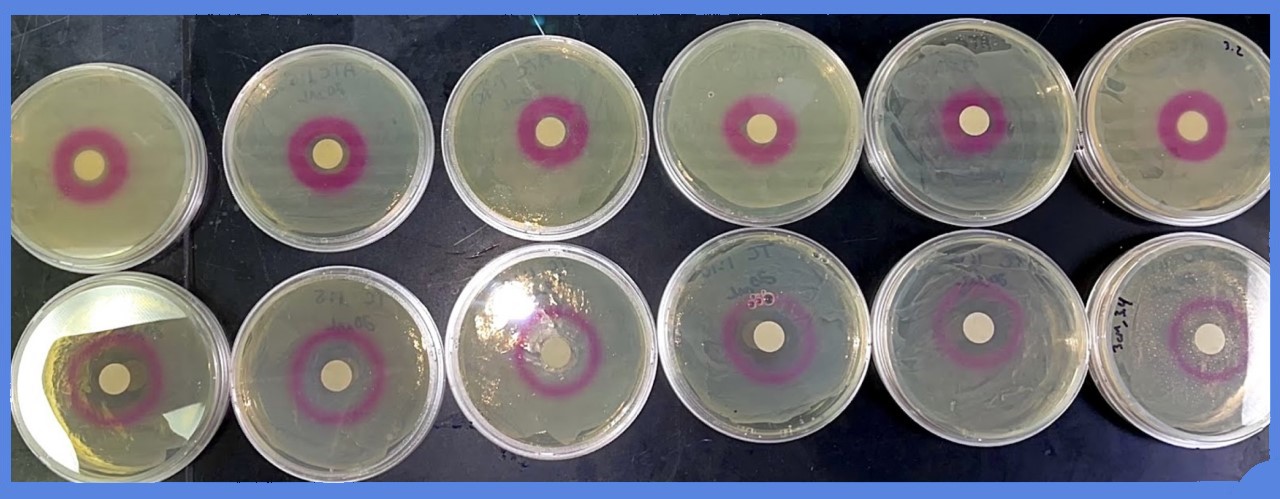
We developed a new assay for measuring induction of this BioBrick, based upon the classic antibiotic filter disc assay.
This assay can be carried out with simple equipment - no fluorimeter is necessary, yet induction of TetR genes can be measured in a quantitative fashion based on diffusion of anhydrotetracycline (aTc) from a filter disc.
Dilutions of aTc (from 5mg/mL to less than 5ug/mL) in EtOH were added to a filter disc, and plated onto an LB agar plate with chloramphenicol (30ug/mL). The plate was previously inoculated with strain HL2483, which constitutively expresses TetR, and the BioBrick I13521.
Representative images are shown below:
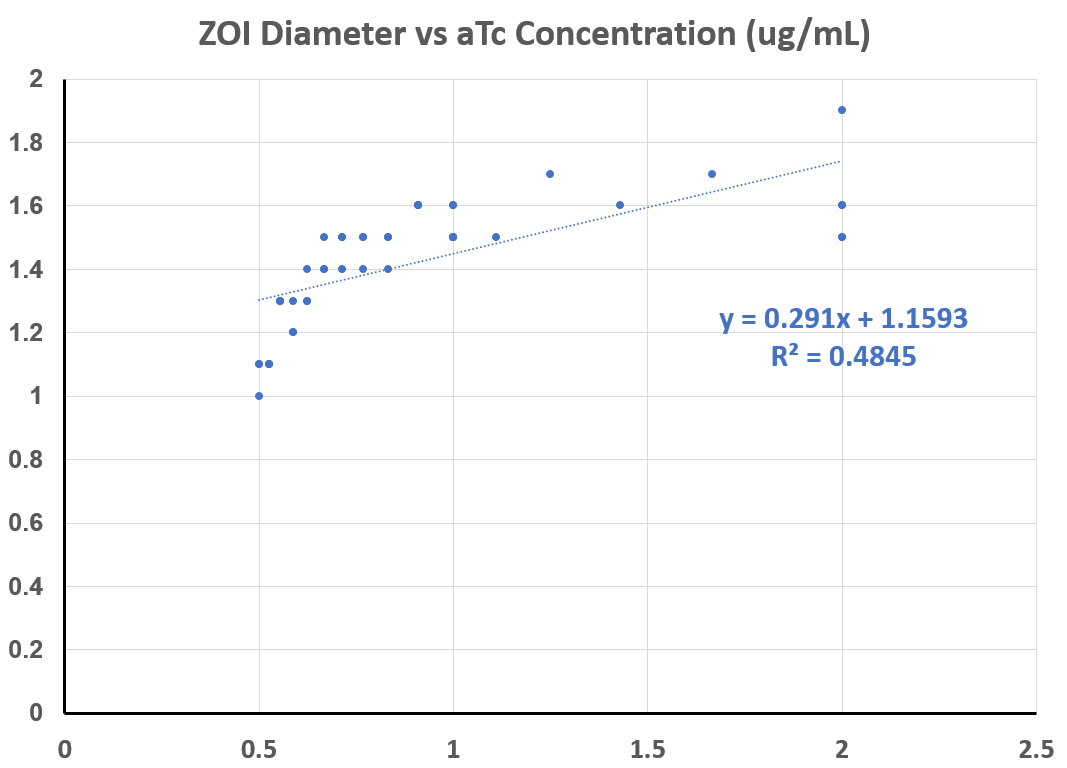
Multiple experiments, at different concentrations of aTC, were carried out for the following quantitative data: the diameter of the zone in which red fluorescent protein was produced was measured - the ZOI, or "zone of induction".
As you can see from the graphs below, and the regression performed, this assay does detect a proportional increase in expression with increasing amounts of inducer.
| emission | RFP |
| excitation | |
| tag | None |

 1 Registry Star
1 Registry Star