This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
RANDOM SEQUENCE FOUND WITHIN PART
CGCTGATAGTGCTAGTGTAGATCGC is found after the RFP stop codon and before the BioBricks suffix. Should not affect transcription or translation of RFP, but good to keep note of it especially in analyzing sequencing results. (KP of siGEM)
- Please note that the above sequence is the old "barcode" sequence added to all of the original CDSs in the early BioBrick part collections. I.e., it's not a random sequence. See https://parts.igem.org/cgi/htdocs/barcodes.cgi for more information (D. Endy).
- FURTHER NOTE The Registry is not displaying barcodes on any of the original parts. The presented sequence information is wrong. This is a serious bug in the Registry that need to be fixed (D. Endy). Drew 14:34, 1 November 2010 (UTC)
Applications of BBa_E1010
User Reviews
UNIQ1d17963ce2a57d16-partinfo-00000000-QINU
|
••••
KAIST_iGEM_2012
|
 Figure 1. E.coli strain MG1655 expressing BBa_E1010 in LB under control of BBa_K907005
BBa_E1010 was successfully used to produce mRFP in E.coli strain MG1655 in LB or M9 minimal media under the control of promoter-BBa_J23119 and RBS-BBa_B0034 in the Dual Phase Protein Generator(mRFP default), BBa_K907005
The expression is clearly observed with naked eye after overnight culture.
|
;
|
•••
DTU_igem_2010
|
Characterization of RFP BBa_E1010
We have characterized RFP BBa_E1010 in two different chassis to test the compatibility and the possible range of expressions before limitations in the cell metabolism.
Method
We have made constructs with a synthetic promoter library (SPL) in front of the E1010, by using BBa_I13507 and the plasmid backbone pSB3T5. For information on design of an SPL compatible with the BB standard see [http://bbf.openwetware.org/RFC.html#BBF_RFC_63:_DTU_Synthetic_Promoter_Library_Standard BBF RFC63].
We have benchmarked the relative promoter strength range achieved from the SPL to the standard promoter BBa_J23101, by calculating the relative promoter strength in vivo as suggested in [http://bbf.openwetware.org/RFC.html#BBF_RFC_19:_Measuring_the_Activity_of_BioBrick.E2.84.A2_Promoters_Using_an_In_Vivo_Reference_Standard BBF RFC 19]. For further explanation on methods see our [http://2010.igem.org/Team:DTU-Denmark iGEM_DTU_2010 wiki].
Results
We show that the RFP E1010 can be expressed with the following results
- In XL1blue with an RPU range form 0 to at least 1,13 RPU.
- In DHA5&alpha with an RPU range from 0 to 1,35 RPU.
 Graph2 XL1BLUE illustrates the variation in promoter strengths of the SPL mapped against the reference promoters. PM corresponds to BBa_J23101, PS corresponds to BBa_J23100 and PW corresponds to BBa_J23116.  Graph3 XL1BLUEillustrates the specific activities of the SPL promoters ranked together with the reference promoters. 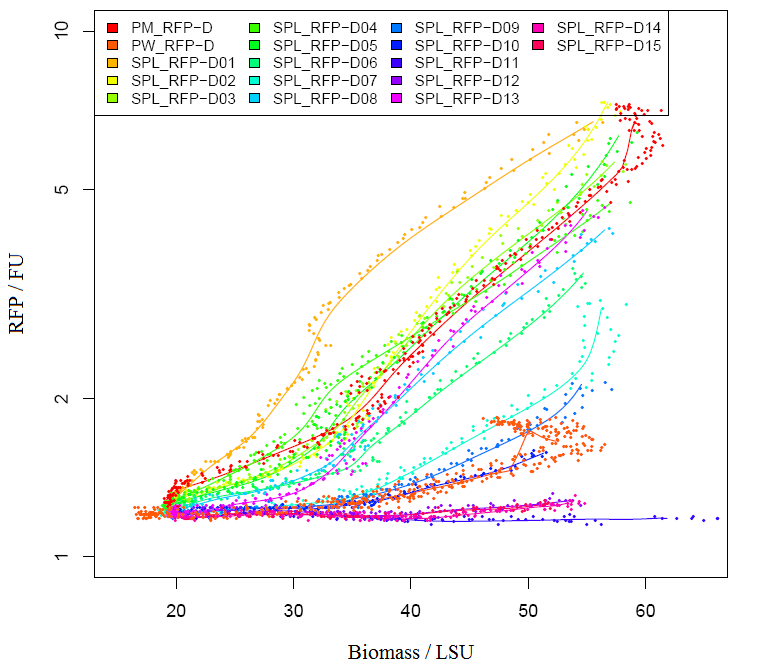 Graph4 DH5aillustrates the variation in promoter strengths of the SPL mapped against the reference promoters. PM corresponds to BBa_J23101, PS corresponds to BBa_J23100 and PW corresponds to BBa_J23116. 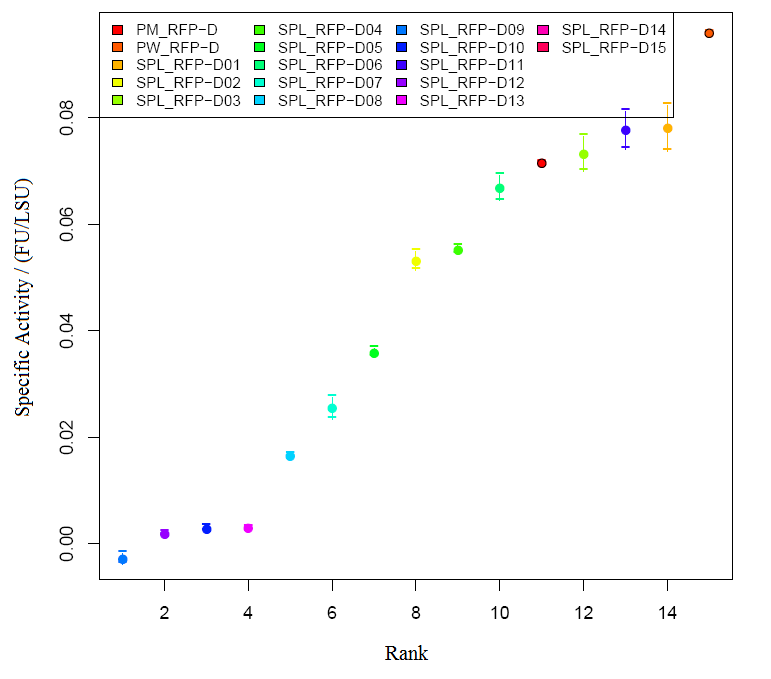 Graph5 DH5a illustrates the specific activities of the SPL promoters ranked against the reference promoters.
Table 1 shows the specific activities and RPUs calculated for all the SPL constructs run in BioLector in XL1-blue
Construct | Specific Activity | RPU |
| BBa_J23101 | 0.0795 | 1.00 |
| SPL_RFP01 | 0.00578 | 0.0727 |
| SPL_RFP02 | 0.0418 | 0.526 |
| SPL_RFP03 | 0.0612 | 0.770 |
| SPL_RFP04 | -0.00027 | -0.00340 |
| SPL_RFP05 | 0.0418 | 0.526 |
| SPL_RFP06 | 0.0856 | 1.08 |
| SPL_RFP07 | 0.0134 | 0.168 |
| SPL_RFP08 | 0.0534 | 0.672 |
| SPL_RFP09 | 0.0638 | 0.803 |
| SPL_RFP10 | 0.00260 | 0.0327 |
| SPL_RFP11 | 0.0900 | 1.13 |
| SPL_RFP12 | 0.0600 | 0.755 |
| SPL_RFP13 | 0.0754 | 0.949 |
| SPL_RFP14 | 0.00795 | 0.100 |
| SPL_RFP16 | 0.00959 | 0.121 |
Table 2 shows the specific activities and RPUs calculated for all the SPL constructs run in BioLector in DH5alpha
Construct | Specific Activity | RPU |
| BBa_J23101 | 0.0712 | 1.00 |
| SPL_RFP-D01 | 0.0715 | 1.00 |
| SPL_RFP-D02 | 0.0959 | 1.35 |
| SPL_RFPD-03 | 0.0781 | 1.10 |
| SPL_RFPD-04 | 0.0531 | 0.746 |
| SPL_RFPD-05 | 0.0732 | 1.03 |
| SPL_RFPD-06 | 0.0552 | 0.775 |
| SPL_RFPD-07 | 0.0358 | 0.503 |
| SPL_RFPD-08 | 0.0668 | 0.938 |
| SPL_RFPD-09 | 0.0254 | 0.357 |
| SPL_RFPD-10 | 0.0165 | 0.231 |
| SPL_RFPD-11 | -0.00284 | -0.399 |
| SPL_RFPD-12 | 0.00276 | 0.0387 |
| SPL_RFPD-13 | 0.0776 | 1.09 |
| SPL_RFPD-14 | 0.0018 | 0.0253 |
| SPL_RFPD-15 | 0.00302 | 0.0424 |
|
;
|
Antiquity
|
This review comes from the old result system and indicates that this part did not work in some test.
|
|
No review score entered.
Nkessler
|
We successfully used this part for a read out system, e.g. in BBa_K389016. Additionally we compared it with a luciferase: BBa_K389004.
|
UNIQ1d17963ce2a57d16-partinfo-0000000C-QINU


 1 Registry Star
1 Registry Star


