Difference between revisions of "Part:BBa K1073022"
| Line 33: | Line 33: | ||
<b>Absorbance spectrum:</b> | <b>Absorbance spectrum:</b> | ||
| − | Below is an absorbance spectrum for | + | Below is an absorbance spectrum for eforRed between 400nm and 850nm. Peak maxima is seen around 580nm. |
<center>https://static.igem.org/mediawiki/2015/4/40/EforRed_spectrum.png</center> | <center>https://static.igem.org/mediawiki/2015/4/40/EforRed_spectrum.png</center> | ||
Revision as of 01:20, 19 September 2015
Constitutively expressed chromoprotein eforRed
This device is an enhanced construct of 2012 Uppsala's BBa_K592012. It consists of a strong promoter (BBa_J23100) combined with a RBS (BBa_B0032) and the encoding region of the chromoprotein eforRed (BBa_K592012). This results in a constitutively high expression of eforRed.
Usage and Biology
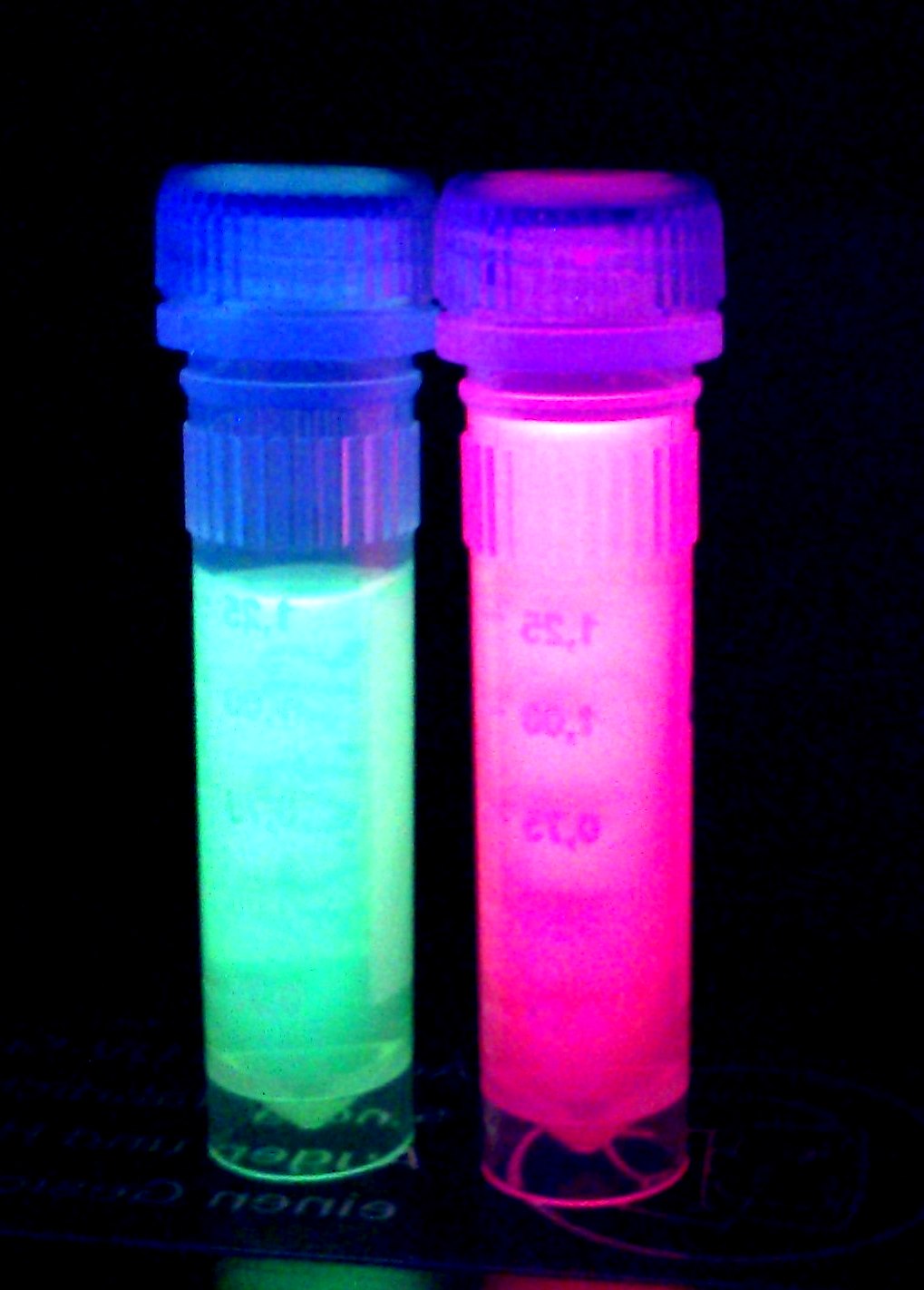
During cultivation, the chromoprotein eforRed is expressed and stored intracellularly. The pink color of the protein can be seen by the naked eye, thus the chromoprotein can be used as a reporter to distinguish cells easily.
iGEM Team Braunschweig 2013:
Left: Liquid cultures of E. coli XL1 expressing chromoproteins amilGFP (BBa_K1073024), eforRed and aeBlue (BBa_K1073020).
Right: Cell pellet of E. coli XL1 expressing eforRed.
The pink color of eforRed can be seen by the naked eye. The chromoprotein also fluoresce when excited. Thus, it can be used to identify cells with fluorescence microscopy or flow cytometry. For further information as well as absorption and emission spectra see experience page.
iGEM Team Braunschweig 2013: Supernatant after cell disruption containing chromoproteins of eforRed and amilGFP (BBa_K1073024). Both chromoproteins fluoresce when excited by UV-light.
Exeter iGEM 2015:
As a part of our project, three chromoproteins BBa_K1431814, BBa_K1073022, and BBa_K1073020 were characterised as they could be used as potential indicators in our cell-free diagnostic test.
eforRed was further characterised in three main ways; an absorbance spectrum was recorded, a standard curve of protein concentration vs. OD458 was calculated, and a visual limit was determined.
Absorbance spectrum: Below is an absorbance spectrum for eforRed between 400nm and 850nm. Peak maxima is seen around 580nm.

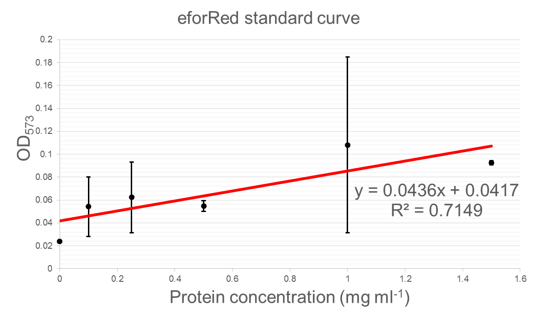
Standard curve - protein concentration vs. OD: Below is a standard curve for concentration of protein vs. OD (573 nm). This curve can be used to determine the concentration of eforRed based on its optical density (OD).
b>Visual limit:</b> Below is an image showing the intensity of colour at a specific amount of chromoprotein, from not visible to readily visible. The point of this characterisation is so that the level of protein expression/production required before a visual output in observed can be deduced.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]